Gene
KWMTBOMO14400 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000117
Annotation
PREDICTED:_uncharacterized_protein_LOC101744111_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.165
Sequence
CDS
ATGAGTGTTTTAATATTGTTTTTTGTGTACGTTTTTGTGCCGTGTGTTTTTGGCCAAGAGGCCGCGGCTTGTTATGGTGCTGGCAGCGTTGCAGGTGCAGCAATAGGTGCTTTCATAGCTGCACTTATTTTGGTTGCAGCTGCTTATTACCTTCGCAAATTGTATTGGAAGTCACGTAAAGGCAAGCATCTGGTCTTCTCAACGGATCCGGAATCGGTGAAGGATGAATTCGCGTTTGACAACCCTGGCTTCAAGCAAGACGAGCGTTGGCAGCATGACGCCACCCTGCCCGTGGGCGGGAAGCCCTCAGTCCTGCACGCCGAGTTCACTAAGCCAGAACAGAGTAAAGATGACACGTTTCTCCAGCGCGCTCGTATACGTCGAGTGAAGCTTTGGGCACGAGACTTCACGGGTCTCGGTTTCCGCTGTGGAGGAGGCGCCAGAGATGGAGTCCACGTACACTCAGTGCTCAGGAGTGGGCCTGCCGCCGCGGCTAAGTTACAACCAGGCGACAAAATAAAAAGCATCAAAATACAATTGATGGGGACTCCGTTAGAGGACGCTGTCAGCATTTTGTCTCTCGCGTCTCCATACCCTGTGGAGTTGGAGGTGGTGGAAGGTGGCAGAGCGAGCGGAGACGGCAGGACGGTACATCATCCGCTGCTGAAGAGGGCCGGCTCGACTGGAGACGTTACAACGCTCGAGAAAGAAGGTAAGCTATTGCACCCTCCGAAGTCACCGAACACTTCGAATTCGAACAACTCAACACTGGAAACCAAATACGGGAAGAGTGGCATCAAGAAGATATCCGAGAAGATCATCACTTCGACCCTCGAGAGGAACAAGAAGGAGAAACGAGACGGTCTACCCACCACTTTGGAAAGGGAAAACGAGAAGAACAGCAAAATGGCCAAAAGTAGACATTCAGATGTCCCTAGTGGTCTCAGCAAGCCTGAGAGGAACAAAACGAGACACTCGACAGGAGCTGACGTCATAGTGCCGCCAGAAAACGGGACAAGTCAAATCGAACACGCAGAAATGCAGAAGAGAGATTACGACCCCAAAAGGGGAATGAAGTTCGGCATCAGAGTACTCCCGCCCAACGTGCCTGACGACGGAGTTTTGAGGCAGAAGAGCGTCGAAAACGGAAGCGCCACCGTCGAGAAGAGATCCGTCGATGAAATAGACAAACCCGAGCCGCCACGACCAGCGCCGACCGTACAAATCAAACACGAAAACGAAACCGAACACAAAAAGCCAGTCGTGGCTAAGAGAAGAGAGAAAATGGCGCCGCCCATACCCAACGCTAGAGTTAAAACGAACGAAACTGAGAGTATCAGCCACGAGACCACAGTGTCGGACGCGTCACAAGCAAGCTTCGCCAGAAACGATCTAAACTCGAGCGGCATCAAACGCGACGAGAACGGAATCCCCCAGGAGATACCGCAGCAAATGCTCGACGCCGCCAAAGCGGCCAGAACCAACAGAAAGAGTTCCGTTGAAATATTAAACGAAAAAGAAAGACAAGATAACAGGAAAGAAAGGAAAAGCTCTGCAGAAACATTACATGACAAAGAGAAACATGAGCAACAGAGAGACGAGGCCCCTAAGCCCGCGAAAAAGTCAAAAGGGAAAGCCCCGTCTCCTCCGGACGCCGACAAAAACAAAACAGACGACAGTATCTTGGAACGATTGCAGAGAGTCAAGGATTTTCTTAAAAGCGAAAAGTACCATTCAAGCTTACTGAACGACTCTAGTGCGGACACGTCAGGTCGAGCGAGTGCCACACGGATCAACACTTCCACGCCAAAGGTCAACAAAACGAAAAGCACTAGCTCCACGCGTCTCGAAGACTCAATCAATTTCAGTCAAGACGACATTGACGATATAGTCTCGAAGCCATTCAAAAGGGAAAGCGACTCACTGAACAACTTCTTCGAGGATTCGAAGTCGAATCTCTCGTCCAACCAGGACGTCCATTCGGTCGTTTCCTTGAACCAGAGCGACAAAAGCGAACGAGGCTCCACGACGATCGAACTAAACAACAGCGACATCACAATACACAGTTCTCCATTGAACGCGACTGTACAGTCCGGGTCCGGCGATACGTCTCTTTTGGATGAAAACGAGAGGAAGGCCGCTTCCCTGGGTGATTTATCCAGGTTCGATTTAAGAACGAACGCCGGTAAACCATCAACCGGTACATTGGAAAGGGCTCAAAGTCTAGACATAACTGCCGACGACGCTGAGATAGCCGAGGCCACGATCTCGCCCAAAAAGAGAAAAGCTATGTCTGTGGTCGAAACTAACTTTTACGAAGCCAGCGGCGAAGACACTTTACCCGACACTCTTGACTCGGACAATGGAATCGTGATAAAACACAAAGAGCCCAGATTGAGTTTGAACGTGAAAACGTCCGCCATGGAAGGCCTTAGCACATTCCAGCGGAATCGTCTCAAGAAAGCATCCGAATTCGGTAACTTGGAGGACGCTATCGTCAAAGGATCGAGCAGCTCTATAGAATCGGACAAAGTCGACGAAGATCACGACAAGAGAACCAAATCCGGACTGCGTATTGGAAGCGAACACGAGACATCCGATCATTTAGCCAAGAGAATCATGGACGAGAACATGAAAGTCCATTTGAAGCTTGTATCGGAATTCGCTAAGACCACTTCAGACGGTAGCACCCTGGCACCACCAGACAACGATCCGCCGACACAACCTAACCTTACGAAACTCGAAACGACACCGACCAAATACGATGAGAAAATAACGATGAACTACGACACAAATATACCGGACGACTTAAAAGTATCGCGCAGTTCGTACGCGAACAGCTTGGAGAGACCAAAATCCGAGATGATGAAGAAACTGCTCGCCAAAAATCCTATACTCAACGTACACATCGACCAGAGCACGCAAAGAGAGGCTTCTACCAGTGCCGAAGTCCAAACGCCGATGACTGACGCTTCGAAATCCGCCATGCACCAACCCGACATAGTCAACTTCGATCTGAAACCCTCCGAAATGAAAACTAAAGATTACGAGGACTTCGTCAGCAACATCAGGGTCGGCTCGAACAATAGCAGCTTGAATAAGAAAATGGAAAGCTTCTCCACCGACTGGCGCGATTCAAAGGAGGGCAGCGATCTCGTCACATTCACCACAGACAGAACTCAAAATCTTGAAGACAAGTCACGTAAGACGTACACGAAATCGATTGAAATCGGCGAACCGACAATGAGGGTAATGCCGCCTGATGTCCTGGAAGGGATTCGAAGACGGCACGACGAGAAAGAGAACCGGACGCTTTACATGGAACCGGGCAACGTATCCCTGACAATGACACAAGAACCGACCCAAAAAACCGTCACAGTCAATGTAGCCGAAGACGAATTCGGGAACAAAGTCATAACGCGAAACGTCGAACAAATATCCACTAAATACGTCACCGCAAAAACTGAAGCACCTCTCCAAGTCGAGCAGTTCAGTTTTGGCATCATGAGAAACGCCGGTCCAACAGACGAACTGGCTCTAGTGGACGGCAACGTTAAGAATATCGATAAGAACGTACTGGACGAAATAAGACGGCAGAACCCGAATATGCATTTCGCATCGGACGAACCTTCGTACACCAAAACGGAGACAATAATTCTGAGCACTTCAGACATGGACGAGGGCCAAGCGAGGGCGTTGATGGAGAAAATGAAGAATGACCCTAGTTTCGTGAGGCGCAAAACGCCAGAGGAACCTACGCGATTAGGTGTCAGGATAACGCAGGATCTGGAAGACGTACAAACGTTCACTGAAGACGCTACCGAACTCACCAAGACCAGATACACCATCCACCCAGCATCAGCGTCCGACGAACAATGCAAATTAGAATCTCACGACGACGCGCACGTAACCGAGATAGAAGTGGTCGCAACGCGCGCCGACAGAAACGCTAAAGGGAAACCGGATAAGGTATCAGGCAGACAAAGGGTCCCGTATCGAGAACCGGTTCTATCGTACGAACTTGACATCGAGCTGTTGAGCAATTTTATATCGAACGAGAGATACCACTCGGCGAGACAGATGGCGGAGGCCAAGAAAACCAAAACGACCGCCTCAGACCCAAAGAAACGCCACTCCGACTACGACTTGCCCAGAAACAGCCATATCAAGTTCAGAACGGCGACGTACGAATCTCCAAAGGGCACGATAGTGACGAGCACGGATCTGGAAAACCGGAGACTATCGGAGCTAGATCAGATGCAGCTGAGATCGGTCAGTCCGCCCCAAAAACCGGTGTTGCTCGCGAAACCGAGCAGCATACCGGTGAAGGCAACCGAGAAAACCGCCAAAACCGGCGTCTCGTCGAAGATCCCGATCGTCTCGGCGCTTAAGTCCTCCAGCCAGGAGAACTTGGCTGAGAGTAGATTCTCTTTGCCGCGATCCCCCCCTCCCCAATACGGGAGCTCCGGTAACATATCTGTAACTTCGATCAAGAGCAGCTCTAGAAGTCCGAGCGGCGGGAAATTTTAA
Protein
MSVLILFFVYVFVPCVFGQEAAACYGAGSVAGAAIGAFIAALILVAAAYYLRKLYWKSRKGKHLVFSTDPESVKDEFAFDNPGFKQDERWQHDATLPVGGKPSVLHAEFTKPEQSKDDTFLQRARIRRVKLWARDFTGLGFRCGGGARDGVHVHSVLRSGPAAAAKLQPGDKIKSIKIQLMGTPLEDAVSILSLASPYPVELEVVEGGRASGDGRTVHHPLLKRAGSTGDVTTLEKEGKLLHPPKSPNTSNSNNSTLETKYGKSGIKKISEKIITSTLERNKKEKRDGLPTTLERENEKNSKMAKSRHSDVPSGLSKPERNKTRHSTGADVIVPPENGTSQIEHAEMQKRDYDPKRGMKFGIRVLPPNVPDDGVLRQKSVENGSATVEKRSVDEIDKPEPPRPAPTVQIKHENETEHKKPVVAKRREKMAPPIPNARVKTNETESISHETTVSDASQASFARNDLNSSGIKRDENGIPQEIPQQMLDAAKAARTNRKSSVEILNEKERQDNRKERKSSAETLHDKEKHEQQRDEAPKPAKKSKGKAPSPPDADKNKTDDSILERLQRVKDFLKSEKYHSSLLNDSSADTSGRASATRINTSTPKVNKTKSTSSTRLEDSINFSQDDIDDIVSKPFKRESDSLNNFFEDSKSNLSSNQDVHSVVSLNQSDKSERGSTTIELNNSDITIHSSPLNATVQSGSGDTSLLDENERKAASLGDLSRFDLRTNAGKPSTGTLERAQSLDITADDAEIAEATISPKKRKAMSVVETNFYEASGEDTLPDTLDSDNGIVIKHKEPRLSLNVKTSAMEGLSTFQRNRLKKASEFGNLEDAIVKGSSSSIESDKVDEDHDKRTKSGLRIGSEHETSDHLAKRIMDENMKVHLKLVSEFAKTTSDGSTLAPPDNDPPTQPNLTKLETTPTKYDEKITMNYDTNIPDDLKVSRSSYANSLERPKSEMMKKLLAKNPILNVHIDQSTQREASTSAEVQTPMTDASKSAMHQPDIVNFDLKPSEMKTKDYEDFVSNIRVGSNNSSLNKKMESFSTDWRDSKEGSDLVTFTTDRTQNLEDKSRKTYTKSIEIGEPTMRVMPPDVLEGIRRRHDEKENRTLYMEPGNVSLTMTQEPTQKTVTVNVAEDEFGNKVITRNVEQISTKYVTAKTEAPLQVEQFSFGIMRNAGPTDELALVDGNVKNIDKNVLDEIRRQNPNMHFASDEPSYTKTETIILSTSDMDEGQARALMEKMKNDPSFVRRKTPEEPTRLGVRITQDLEDVQTFTEDATELTKTRYTIHPASASDEQCKLESHDDAHVTEIEVVATRADRNAKGKPDKVSGRQRVPYREPVLSYELDIELLSNFISNERYHSARQMAEAKKTKTTASDPKKRHSDYDLPRNSHIKFRTATYESPKGTIVTSTDLENRRLSELDQMQLRSVSPPQKPVLLAKPSSIPVKATEKTAKTGVSSKIPIVSALKSSSQENLAESRFSLPRSPPPQYGSSGNISVTSIKSSSRSPSGGKF
Summary
Uniprot
EMBL
Proteomes
PRIDE
SUPFAM
SSF50156
SSF50156
ProteinModelPortal
PDB
4CMZ
E-value=0.0410189,
Score=92
Ontologies
GO
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.676285
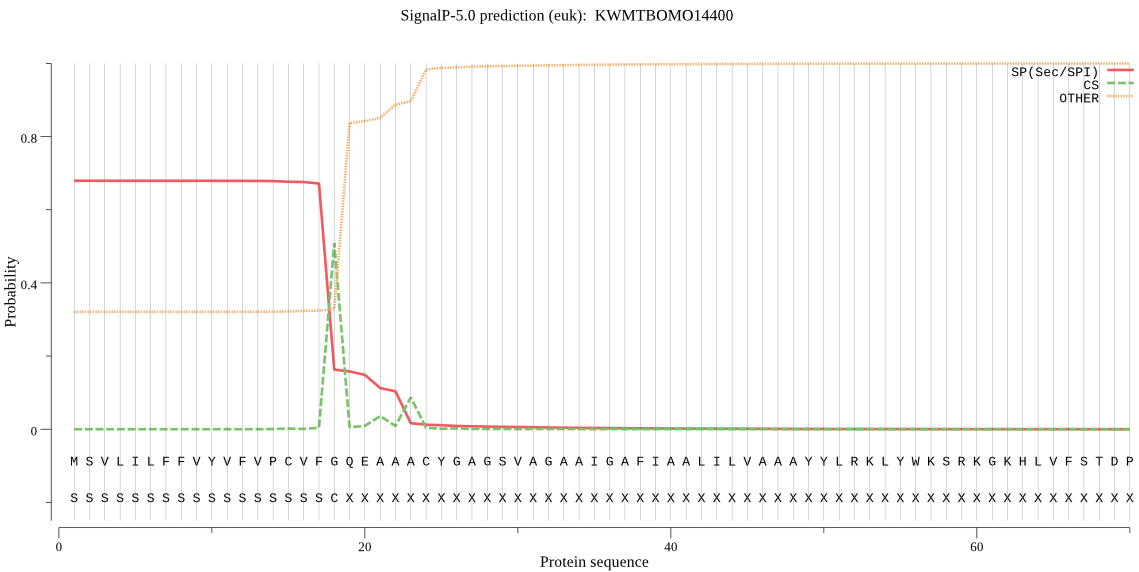
Length:
1509
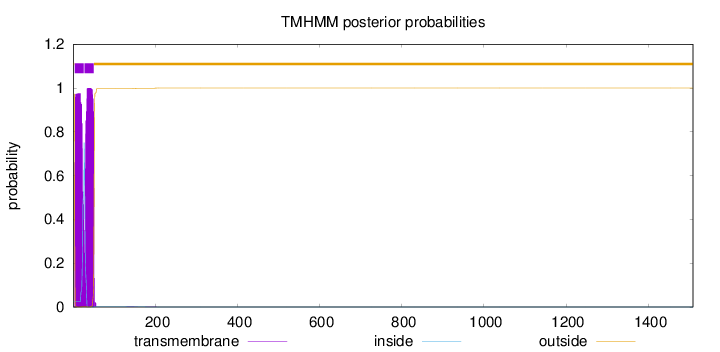
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
42.4516300000001
Exp number, first 60 AAs:
42.39694
Total prob of N-in:
0.02723
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 26
inside
27 - 27
TMhelix
28 - 50
outside
51 - 1509
Population Genetic Test Statistics
Pi
225.061206
Theta
157.798958
Tajima's D
1.124114
CLR
0.196493
CSRT
0.692465376731163
Interpretation
Uncertain
