Gene
KWMTBOMO14390
Pre Gene Modal
BGIBMGA014500
Annotation
PREDICTED:_sperm_surface_protein_Sp17-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.565 Nuclear Reliability : 2.037
Sequence
CDS
ATGAGCTGGCGCCGAATCTTGTTGGAATACCCAGTGCCTGTTATTGAACATGAACTGTGCCACGCCGCAACCAAGATCCAGGCTTCGTTCCGAGGCCACCAGGCGCGGAAGGAGACTCAAGCCCAAAAGGAAAAGGAAAAGCAAGATAAGGAGGACATCGAGAAAATAGATCTGACCGATCCTGATTTGAACAAAGCCGCGACGAAGATCCAGGCCTCATTCCGCGGTCACAAAGTGCGCAAGGACGTCCCGAACTAA
Protein
MSWRRILLEYPVPVIEHELCHAATKIQASFRGHQARKETQAQKEKEKQDKEDIEKIDLTDPDLNKAATKIQASFRGHKVRKDVPN
Summary
Uniprot
H9JY80
A0A194PSK1
A0A2H1WM24
A0A1B6LJH0
A0A0K8SL04
A0A1B6JBH1
+ More
A0A1B6FS87 D6WES8 N6UV16 A0A0A9ZFP6 A0A3Q0IMV4 A0A0K8SJZ7 A0A182HG24 B4J601 A0A1B6E337 A0A2R7VVW7 A0A0N8P008 A0A0Q9WF71 A0A0R3NLU6 A0A1B0DE65 A0A0M3QUV4 A0A0R1DWK7 A0A0C9S2P5 A0A1W4W0Y6 A0A0J9RE98 A0A0Q5VNN9 Q95T29 A0A224XP31 A0A182P348 A0A0C9QK51 K7IXJ3 A0A182U5F4 A0A2M3ZDZ9 A0A182T4K9 A0A182YE82 A0A0Q9XL72 A0A182N339 A0A0R3NM67 Q290Z3 B4GB11 A0A151I835 B4LLM0 B3MGK9 A0A3B0JLB7 A0A182ILR5 A0A1W4WCY9 B4QGD9 B3NQN4 Q7JPS2 B4P7T8 B4HRT9 A0A0J9U2E9 A0A1A9Z5E5 A0A1B0G6I4 A0A2A3E8C0 V9IL74 A0A1A9VVM3 A0A182Q1K0 A0A0Q9WPQ6 B4KP21 A0A154P8M3 A0A0K8UQH9 A0A034V512 A0A1A9XHD8 A0A1B0C223 A0A182LW40 A0A0A1XRC1 E2AKK1 A0A195BIY6 A0A158NJX1 A0A1A9W7T4 W8AIW7 A0A0L0CMF3 F4WIM0 A0A0K8VLF6 A0A195DV53 T1PFS4 A0A026WQ50 A0A195F556 A0A0A1X783 A0A0J7L9V1 A0A151XE82 A0A1B0GKN4 A0A1I8P4T1 E0VBC9 E2BWY7 A0A1I8NDK3 A0A2S2PZ50 A0A2S2NB03 A0A1W4XL40 A0A336KQM8 A0A1W4XWK7 T1JQ30 A0A0P6BP48 J9LCR1 B3RU82
A0A1B6FS87 D6WES8 N6UV16 A0A0A9ZFP6 A0A3Q0IMV4 A0A0K8SJZ7 A0A182HG24 B4J601 A0A1B6E337 A0A2R7VVW7 A0A0N8P008 A0A0Q9WF71 A0A0R3NLU6 A0A1B0DE65 A0A0M3QUV4 A0A0R1DWK7 A0A0C9S2P5 A0A1W4W0Y6 A0A0J9RE98 A0A0Q5VNN9 Q95T29 A0A224XP31 A0A182P348 A0A0C9QK51 K7IXJ3 A0A182U5F4 A0A2M3ZDZ9 A0A182T4K9 A0A182YE82 A0A0Q9XL72 A0A182N339 A0A0R3NM67 Q290Z3 B4GB11 A0A151I835 B4LLM0 B3MGK9 A0A3B0JLB7 A0A182ILR5 A0A1W4WCY9 B4QGD9 B3NQN4 Q7JPS2 B4P7T8 B4HRT9 A0A0J9U2E9 A0A1A9Z5E5 A0A1B0G6I4 A0A2A3E8C0 V9IL74 A0A1A9VVM3 A0A182Q1K0 A0A0Q9WPQ6 B4KP21 A0A154P8M3 A0A0K8UQH9 A0A034V512 A0A1A9XHD8 A0A1B0C223 A0A182LW40 A0A0A1XRC1 E2AKK1 A0A195BIY6 A0A158NJX1 A0A1A9W7T4 W8AIW7 A0A0L0CMF3 F4WIM0 A0A0K8VLF6 A0A195DV53 T1PFS4 A0A026WQ50 A0A195F556 A0A0A1X783 A0A0J7L9V1 A0A151XE82 A0A1B0GKN4 A0A1I8P4T1 E0VBC9 E2BWY7 A0A1I8NDK3 A0A2S2PZ50 A0A2S2NB03 A0A1W4XL40 A0A336KQM8 A0A1W4XWK7 T1JQ30 A0A0P6BP48 J9LCR1 B3RU82
Pubmed
19121390
26354079
18362917
19820115
23537049
25401762
+ More
17994087 15632085 17550304 22936249 18057021 7925024 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 25244985 25348373 25830018 20798317 21347285 24495485 26108605 21719571 25315136 24508170 30249741 20566863 18719581
17994087 15632085 17550304 22936249 18057021 7925024 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 25244985 25348373 25830018 20798317 21347285 24495485 26108605 21719571 25315136 24508170 30249741 20566863 18719581
EMBL
BABH01042720
KQ459601
KPI94100.1
ODYU01009593
SOQ54131.1
GEBQ01016125
+ More
JAT23852.1 GBRD01012344 JAG53480.1 GECU01011182 JAS96524.1 GECZ01016719 JAS53050.1 KQ971326 EFA00894.1 APGK01010679 APGK01030326 KB740770 KB738267 KB632132 ENN78987.1 ENN82692.1 ERL88990.1 GBHO01001326 GBHO01001322 JAG42278.1 JAG42282.1 GBRD01012345 JAG53479.1 APCN01005461 APCN01005462 CH916367 EDW00844.1 GEDC01028044 GEDC01004960 JAS09254.1 JAS32338.1 KK854111 PTY11479.1 CH902619 KPU75804.1 CH940648 KRF79659.1 CM000071 KRT01969.1 AJVK01014749 AJVK01014750 CP012524 ALC41319.1 CM000158 KRJ99632.1 GBYB01015176 JAG84943.1 CM002911 KMY93909.1 CH954179 KQS62572.1 KQS62573.1 S72579 AY060354 AE013599 KX531440 AAB32066.1 AAL25393.1 AAM68523.1 AGB93520.1 ANY27250.1 GFTR01002181 JAW14245.1 GBYB01015178 JAG84945.1 AAZX01004493 AAZX01009565 AAZX01011944 GGFM01005909 MBW26660.1 CH933808 KRG05047.1 KRT01970.1 EAL25219.1 CH479181 EDW32113.1 KQ978390 KYM94304.1 EDW60883.1 EDV35752.1 OUUW01000001 SPP74289.1 CM000362 EDX07177.1 EDV55944.1 S72573 AAB32065.1 AAF58168.2 EDW91114.1 CH480816 EDW47953.1 KMY93910.1 CCAG010012808 KZ288328 PBC27940.1 JR050300 AEY61226.1 AXCN02001131 CH963848 KRF97862.1 EDW10087.1 KQ434845 KZC08207.1 GDHF01028849 GDHF01023571 JAI23465.1 JAI28743.1 GAKP01021343 GAKP01021342 JAC37609.1 JXJN01024301 AXCM01018236 GBXI01001169 JAD13123.1 GL440281 EFN66052.1 KQ976465 KYM84299.1 ADTU01018456 GAMC01020893 GAMC01020892 JAB85662.1 JRES01000195 KNC33387.1 GL888176 EGI65956.1 GDHF01021540 GDHF01012605 JAI30774.1 JAI39709.1 KQ980304 KYN16790.1 KA647667 AFP62296.1 KK107135 QOIP01000006 EZA58093.1 RLU21901.1 KQ981820 KYN35307.1 GBXI01007305 JAD06987.1 LBMM01000172 KMR04628.1 KQ982254 KYQ58630.1 AJWK01030758 AJWK01030759 AJWK01030760 DS235026 EEB10685.1 GL451188 EFN79851.1 GGMS01001508 MBY70711.1 GGMR01001533 MBY14152.1 UFQS01000833 UFQT01000833 SSX07124.1 SSX27467.1 CAEY01000437 GDIP01012209 JAM91506.1 ABLF02025006 DS985244 EDV25293.1
JAT23852.1 GBRD01012344 JAG53480.1 GECU01011182 JAS96524.1 GECZ01016719 JAS53050.1 KQ971326 EFA00894.1 APGK01010679 APGK01030326 KB740770 KB738267 KB632132 ENN78987.1 ENN82692.1 ERL88990.1 GBHO01001326 GBHO01001322 JAG42278.1 JAG42282.1 GBRD01012345 JAG53479.1 APCN01005461 APCN01005462 CH916367 EDW00844.1 GEDC01028044 GEDC01004960 JAS09254.1 JAS32338.1 KK854111 PTY11479.1 CH902619 KPU75804.1 CH940648 KRF79659.1 CM000071 KRT01969.1 AJVK01014749 AJVK01014750 CP012524 ALC41319.1 CM000158 KRJ99632.1 GBYB01015176 JAG84943.1 CM002911 KMY93909.1 CH954179 KQS62572.1 KQS62573.1 S72579 AY060354 AE013599 KX531440 AAB32066.1 AAL25393.1 AAM68523.1 AGB93520.1 ANY27250.1 GFTR01002181 JAW14245.1 GBYB01015178 JAG84945.1 AAZX01004493 AAZX01009565 AAZX01011944 GGFM01005909 MBW26660.1 CH933808 KRG05047.1 KRT01970.1 EAL25219.1 CH479181 EDW32113.1 KQ978390 KYM94304.1 EDW60883.1 EDV35752.1 OUUW01000001 SPP74289.1 CM000362 EDX07177.1 EDV55944.1 S72573 AAB32065.1 AAF58168.2 EDW91114.1 CH480816 EDW47953.1 KMY93910.1 CCAG010012808 KZ288328 PBC27940.1 JR050300 AEY61226.1 AXCN02001131 CH963848 KRF97862.1 EDW10087.1 KQ434845 KZC08207.1 GDHF01028849 GDHF01023571 JAI23465.1 JAI28743.1 GAKP01021343 GAKP01021342 JAC37609.1 JXJN01024301 AXCM01018236 GBXI01001169 JAD13123.1 GL440281 EFN66052.1 KQ976465 KYM84299.1 ADTU01018456 GAMC01020893 GAMC01020892 JAB85662.1 JRES01000195 KNC33387.1 GL888176 EGI65956.1 GDHF01021540 GDHF01012605 JAI30774.1 JAI39709.1 KQ980304 KYN16790.1 KA647667 AFP62296.1 KK107135 QOIP01000006 EZA58093.1 RLU21901.1 KQ981820 KYN35307.1 GBXI01007305 JAD06987.1 LBMM01000172 KMR04628.1 KQ982254 KYQ58630.1 AJWK01030758 AJWK01030759 AJWK01030760 DS235026 EEB10685.1 GL451188 EFN79851.1 GGMS01001508 MBY70711.1 GGMR01001533 MBY14152.1 UFQS01000833 UFQT01000833 SSX07124.1 SSX27467.1 CAEY01000437 GDIP01012209 JAM91506.1 ABLF02025006 DS985244 EDV25293.1
Proteomes
UP000005204
UP000053268
UP000007266
UP000019118
UP000030742
UP000079169
+ More
UP000075840 UP000001070 UP000007801 UP000008792 UP000001819 UP000092462 UP000092553 UP000002282 UP000192221 UP000008711 UP000000803 UP000075885 UP000002358 UP000075902 UP000075901 UP000076408 UP000009192 UP000075884 UP000008744 UP000078542 UP000268350 UP000075880 UP000000304 UP000001292 UP000092445 UP000092444 UP000242457 UP000078200 UP000075886 UP000007798 UP000076502 UP000092443 UP000092460 UP000075883 UP000000311 UP000078540 UP000005205 UP000091820 UP000037069 UP000007755 UP000078492 UP000095301 UP000053097 UP000279307 UP000078541 UP000036403 UP000075809 UP000092461 UP000095300 UP000009046 UP000008237 UP000192223 UP000015104 UP000007819 UP000009022
UP000075840 UP000001070 UP000007801 UP000008792 UP000001819 UP000092462 UP000092553 UP000002282 UP000192221 UP000008711 UP000000803 UP000075885 UP000002358 UP000075902 UP000075901 UP000076408 UP000009192 UP000075884 UP000008744 UP000078542 UP000268350 UP000075880 UP000000304 UP000001292 UP000092445 UP000092444 UP000242457 UP000078200 UP000075886 UP000007798 UP000076502 UP000092443 UP000092460 UP000075883 UP000000311 UP000078540 UP000005205 UP000091820 UP000037069 UP000007755 UP000078492 UP000095301 UP000053097 UP000279307 UP000078541 UP000036403 UP000075809 UP000092461 UP000095300 UP000009046 UP000008237 UP000192223 UP000015104 UP000007819 UP000009022
Pfam
PF00612 IQ
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JY80
A0A194PSK1
A0A2H1WM24
A0A1B6LJH0
A0A0K8SL04
A0A1B6JBH1
+ More
A0A1B6FS87 D6WES8 N6UV16 A0A0A9ZFP6 A0A3Q0IMV4 A0A0K8SJZ7 A0A182HG24 B4J601 A0A1B6E337 A0A2R7VVW7 A0A0N8P008 A0A0Q9WF71 A0A0R3NLU6 A0A1B0DE65 A0A0M3QUV4 A0A0R1DWK7 A0A0C9S2P5 A0A1W4W0Y6 A0A0J9RE98 A0A0Q5VNN9 Q95T29 A0A224XP31 A0A182P348 A0A0C9QK51 K7IXJ3 A0A182U5F4 A0A2M3ZDZ9 A0A182T4K9 A0A182YE82 A0A0Q9XL72 A0A182N339 A0A0R3NM67 Q290Z3 B4GB11 A0A151I835 B4LLM0 B3MGK9 A0A3B0JLB7 A0A182ILR5 A0A1W4WCY9 B4QGD9 B3NQN4 Q7JPS2 B4P7T8 B4HRT9 A0A0J9U2E9 A0A1A9Z5E5 A0A1B0G6I4 A0A2A3E8C0 V9IL74 A0A1A9VVM3 A0A182Q1K0 A0A0Q9WPQ6 B4KP21 A0A154P8M3 A0A0K8UQH9 A0A034V512 A0A1A9XHD8 A0A1B0C223 A0A182LW40 A0A0A1XRC1 E2AKK1 A0A195BIY6 A0A158NJX1 A0A1A9W7T4 W8AIW7 A0A0L0CMF3 F4WIM0 A0A0K8VLF6 A0A195DV53 T1PFS4 A0A026WQ50 A0A195F556 A0A0A1X783 A0A0J7L9V1 A0A151XE82 A0A1B0GKN4 A0A1I8P4T1 E0VBC9 E2BWY7 A0A1I8NDK3 A0A2S2PZ50 A0A2S2NB03 A0A1W4XL40 A0A336KQM8 A0A1W4XWK7 T1JQ30 A0A0P6BP48 J9LCR1 B3RU82
A0A1B6FS87 D6WES8 N6UV16 A0A0A9ZFP6 A0A3Q0IMV4 A0A0K8SJZ7 A0A182HG24 B4J601 A0A1B6E337 A0A2R7VVW7 A0A0N8P008 A0A0Q9WF71 A0A0R3NLU6 A0A1B0DE65 A0A0M3QUV4 A0A0R1DWK7 A0A0C9S2P5 A0A1W4W0Y6 A0A0J9RE98 A0A0Q5VNN9 Q95T29 A0A224XP31 A0A182P348 A0A0C9QK51 K7IXJ3 A0A182U5F4 A0A2M3ZDZ9 A0A182T4K9 A0A182YE82 A0A0Q9XL72 A0A182N339 A0A0R3NM67 Q290Z3 B4GB11 A0A151I835 B4LLM0 B3MGK9 A0A3B0JLB7 A0A182ILR5 A0A1W4WCY9 B4QGD9 B3NQN4 Q7JPS2 B4P7T8 B4HRT9 A0A0J9U2E9 A0A1A9Z5E5 A0A1B0G6I4 A0A2A3E8C0 V9IL74 A0A1A9VVM3 A0A182Q1K0 A0A0Q9WPQ6 B4KP21 A0A154P8M3 A0A0K8UQH9 A0A034V512 A0A1A9XHD8 A0A1B0C223 A0A182LW40 A0A0A1XRC1 E2AKK1 A0A195BIY6 A0A158NJX1 A0A1A9W7T4 W8AIW7 A0A0L0CMF3 F4WIM0 A0A0K8VLF6 A0A195DV53 T1PFS4 A0A026WQ50 A0A195F556 A0A0A1X783 A0A0J7L9V1 A0A151XE82 A0A1B0GKN4 A0A1I8P4T1 E0VBC9 E2BWY7 A0A1I8NDK3 A0A2S2PZ50 A0A2S2NB03 A0A1W4XL40 A0A336KQM8 A0A1W4XWK7 T1JQ30 A0A0P6BP48 J9LCR1 B3RU82
Ontologies
GO
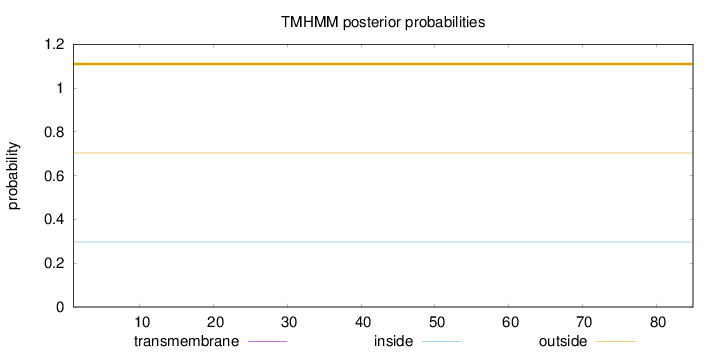
Topology
Length:
85
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.29640
outside
1 - 85
Population Genetic Test Statistics
Pi
209.31598
Theta
59.501116
Tajima's D
2.891319
CLR
0.212566
CSRT
0.972051397430128
Interpretation
Uncertain
