Gene
KWMTBOMO14383
Pre Gene Modal
BGIBMGA013310
Annotation
PREDICTED:_irregular_chiasm_C-roughest_protein-like_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.231 Nuclear Reliability : 1.79 PlasmaMembrane Reliability : 1.103
Sequence
CDS
ATGTCGAACTACAGACCGAGGACAACTAGTAGGGATGTGAAACCGTTGTTTCTGTTGTTGCGGACCAAGTATGTGCGTTTTTACGTGTTCAGTGCGATATTTATCAGTTCGTTCAGTGTTTGTTGCGGCTTCGCGGAGCAGAAGTTCGCCATGGAGCCACAGGATCAGACCGCCGTTATTGGCTCGAGGGTGACACTGCCATGCAGAGTTGAAAACAAGGTGGGTCAACTCCAGTGGACAAAGGATGATTTCGGCCTTGGACTCCACAGAGAGTTGAGCGGCTATGACCGGTATAAGATGATCGGTAGCGACGAGGAAGGTGACTACTCTTTGGACATCAGAGACGTAAATTTAGATGACGACGCCAAATACCAGTGCCAAGTCAGCTCAGGAGTTAAAGGTGAACCAGCAATTAGGTCACGATACGCCCGTCTCACCGTGCTAGTTCCGCCGGAACCACCGAAAATACTGCAAGGGAATTTCTACTCAACCACCGAAGACAGAAATATCAAATTGGAATGCGTGTCGGTCGGCGGAAAGCCGCCCGCTGAGATAACGTGGGTGGATAGCAACGCCGGAGTTTTGACTCAAGGTGTCACTTACACCGTGGAGCCCCTGTCTGACGGACAAAGGTTTACTGCGAGGTCAATTATAAATATGACGCCCAAGAAGGAACATCACAACCAAACTTTCATTTGTCAAGCTCAGAACACAGCGGACCGGGCGTACAGGGCGGCCAGTATACAAATCGAGGTGAAATACGCTCCTAAAGTGAAGATTTACATAAAATCAAGAGTCAATGGGAGAATTCCCGAAGGGGATGATGTAACAATAGGCTGCAAAGCAGACGCCAATCCAACGAATCTAACTTACAAATGGTATATAAATGAGGAATCTATAAATGGAAACACTACTGAATTGAAAATCTACAACATATCAAGAAAGTACCACGACGCAATAGTGAAATGCGTTGTTTACAACGACGTCGGAAAGACTGAGGAAGAGGAGACCTTGGAGATTACTTATGCGCCATCCTTTCGCCATCTCCCCAAGGACGTCGAAGCCGAAGTTGGCACTAGCGTAACCTTGAGTTGCGATGTGGATAGCTACCCGAATCCAGAGATACGATGGCTCCATCATGAAGAAGACCAGACTATTCGCGTGGGACGCACCCCTAATTTAACTCTCACAGTGGATACCCACACATCAGGGAGGTATCTTTGTAAAGCCAGCGTGGACGGATTCCCTGAAATAGAAGCAGAGGCGTCGGTGTTCATTAAAGGTCCTCCGAAGATAATATCGAATCAGACACAATTCGGTACCCAGGGGGACACAGTGAACATTGAGTGTGCAGCCTTCGCCGTGCCGAGGATAGACAGTATTATCTGGACTTTTAACGGAAGAGAAATAGATTCGTTGCATGATCAGGACTACGCATTCCTCGAAGATCTACAATCAGGAGGCATAGTTAACTCTACGCTAATAATTCGAGAGAGCCAATACAAGCACTTCGGCGTGTACAAATGTAACGTGTCTAATGACTATGGCAATGACTATTTGGAGATTACCGTGACTGCTAAAGAGGCCCTTCCACTGCTGGTGATAATTTTCAGCGTAACCACTTGCACGATTGCCGTGGTTTGCCTAACTATGCTGATAATAATTTGCCAAAGAAAGCAAAAAAGGGGAAAGCAGAGTCAAGTCGCCGAGAAGCCAGATGTGACTGTCACGGCCCAAGAGCTTTTCAAGGATAACGACAGGAACTCAAACATTAGCGACCTGAAGCTTGAACTAAGACAGGCCAACGGTAGCTGTGAAATTGATTATTCCAACACCGGCTCAGATAGTACCTTGTGTTCTAATGCCAAATTAGGCAGCGGCATTCCATTGGCCGGTTCTGTTCCTTTAGCATCTATGAACCAGAATAACTATCAGCCGTACAGATATAGTAACGATTACACAGATCCTGCTTACGCTGATTGTTACAAGGTCAATGGCTTTAACAACGGCTATCAGACGTATGTTCATTATGGACATGATTACACGCCTGGTTCGTTGAGGGTGCAGTCGCCGACTTTGGCCAGCGATAAACTCACCCCTAACAGGTCTATAAACGGAAGTTTACCTAGAAGTGTCGATACGCCATCGACGGTTCAATACAACGTAGGGAACGGATCCCAAAATAACTCGTTGCAGCGTTCAACAAAACGGCAAGACAGCAGCAGCGCATTGAATAACCTGGGCCTTCCTACAGCAGAGGTAAGCCGCGTGCCTAATGGTGGCATACATGGTGTTGATATAAGATACGCAGCAACATATGGGAATCCACATTTACGTTCTGGAGGTGGACTGGGTTTCAATACTGTTAACACTGCCAAACCGGCGTCCACTCCAGCGCCACCACCTTATTCGTCTGTCAGAAATTCTGTAATATTACCTTCAGGTAACTCCTCTCACTCTATAACATCGCCAACAAGTGTAGCCTCACCACAATGCGCTACTAGTCCTATAACAACGTCAACTATATCCACTGACAGCACCCAGCCGCTGACCGGCGTGGCAACTTCGTCCGCGACCCCACAGTCTCCTAGCGCACATTACATACTCGCACCATCGAACAGGAGTATAACAACGGCCGCTGTTACTAAGAAAGGTAATGGCTTGCAATCACCCTTGGCAACGCACGTATAA
Protein
MSNYRPRTTSRDVKPLFLLLRTKYVRFYVFSAIFISSFSVCCGFAEQKFAMEPQDQTAVIGSRVTLPCRVENKVGQLQWTKDDFGLGLHRELSGYDRYKMIGSDEEGDYSLDIRDVNLDDDAKYQCQVSSGVKGEPAIRSRYARLTVLVPPEPPKILQGNFYSTTEDRNIKLECVSVGGKPPAEITWVDSNAGVLTQGVTYTVEPLSDGQRFTARSIINMTPKKEHHNQTFICQAQNTADRAYRAASIQIEVKYAPKVKIYIKSRVNGRIPEGDDVTIGCKADANPTNLTYKWYINEESINGNTTELKIYNISRKYHDAIVKCVVYNDVGKTEEEETLEITYAPSFRHLPKDVEAEVGTSVTLSCDVDSYPNPEIRWLHHEEDQTIRVGRTPNLTLTVDTHTSGRYLCKASVDGFPEIEAEASVFIKGPPKIISNQTQFGTQGDTVNIECAAFAVPRIDSIIWTFNGREIDSLHDQDYAFLEDLQSGGIVNSTLIIRESQYKHFGVYKCNVSNDYGNDYLEITVTAKEALPLLVIIFSVTTCTIAVVCLTMLIIICQRKQKRGKQSQVAEKPDVTVTAQELFKDNDRNSNISDLKLELRQANGSCEIDYSNTGSDSTLCSNAKLGSGIPLAGSVPLASMNQNNYQPYRYSNDYTDPAYADCYKVNGFNNGYQTYVHYGHDYTPGSLRVQSPTLASDKLTPNRSINGSLPRSVDTPSTVQYNVGNGSQNNSLQRSTKRQDSSSALNNLGLPTAEVSRVPNGGIHGVDIRYAATYGNPHLRSGGGLGFNTVNTAKPASTPAPPPYSSVRNSVILPSGNSSHSITSPTSVASPQCATSPITTSTISTDSTQPLTGVATSSATPQSPSAHYILAPSNRSITTAAVTKKGNGLQSPLATHV
Summary
Uniprot
A0A2W1BXG4
A0A2A4K6N5
A0A212F6J5
A0A194QIS4
A0A2H1VR82
A0A2A4J512
+ More
A0A2A4J6J8 A0A2A4J5B3 A0A194QH44 A0A0N1IAU7 A0A2W1BS56 A0A212F1H1 A0A2A4K797 D6WHF2 A0A139WKZ8 A0A139WL53 A0A212EGX2 A0A1Y1MDY1 A0A1Y1MGV5 A0A1W4WYV5 A0A1W4WYL8 A0A1B6CCC2 A0A1B6DD38 A0A3S2LLT4 N6UM56 U4UA94 A0A2W1BD20 A0A224XJU8 A0A0A9XJP9 A0A1S4EH93 A0A182GTF8 A0A336LIT5 A0A182XXZ4 Q17HA9 A0A182W5B9 A0A182PL60 A0A1I8PJC8 A0A084VGV9 A0A182RCJ2 A0A182JDB9 A0A1I8NE46 A0A182FIZ2 Q7QEY8 A0A1I8NE48 A0A182VPE7 A0A182U923 B4NDY0 B0WQS2 A0A182J1W2 A0A1B6MEQ7 A0A182PBZ6 A0A0A1XF74 A0A182GAK6 A0A1J1HPX0 A0A1B6LJR2 A0A182RNF1 A0A182QTQ6 A0A182R0F3 A0A182W5C0 W8BE24 A0A3B0K780 A0A182NVF9 A0A1A9V0N2 Q29HP2 A0A0R3P510 A0A1A9ZLV7 A0A182GHE0 A0A1B0FNC9 A0A1A9XDY6 A0A1B0B8K6 A0A0K8VDK1 A0A182IAD7 A0A1A9WGC4 A0A182UYI5 A0A182SR59 A0A182XXZ3 Q9N9Y9 Q9W4T9 A0A1B0CQR5 A0A182IAD8 A0A182M1Q9 A0A182NVG0 A0A182WTP5 A0A182KDT0
A0A2A4J6J8 A0A2A4J5B3 A0A194QH44 A0A0N1IAU7 A0A2W1BS56 A0A212F1H1 A0A2A4K797 D6WHF2 A0A139WKZ8 A0A139WL53 A0A212EGX2 A0A1Y1MDY1 A0A1Y1MGV5 A0A1W4WYV5 A0A1W4WYL8 A0A1B6CCC2 A0A1B6DD38 A0A3S2LLT4 N6UM56 U4UA94 A0A2W1BD20 A0A224XJU8 A0A0A9XJP9 A0A1S4EH93 A0A182GTF8 A0A336LIT5 A0A182XXZ4 Q17HA9 A0A182W5B9 A0A182PL60 A0A1I8PJC8 A0A084VGV9 A0A182RCJ2 A0A182JDB9 A0A1I8NE46 A0A182FIZ2 Q7QEY8 A0A1I8NE48 A0A182VPE7 A0A182U923 B4NDY0 B0WQS2 A0A182J1W2 A0A1B6MEQ7 A0A182PBZ6 A0A0A1XF74 A0A182GAK6 A0A1J1HPX0 A0A1B6LJR2 A0A182RNF1 A0A182QTQ6 A0A182R0F3 A0A182W5C0 W8BE24 A0A3B0K780 A0A182NVF9 A0A1A9V0N2 Q29HP2 A0A0R3P510 A0A1A9ZLV7 A0A182GHE0 A0A1B0FNC9 A0A1A9XDY6 A0A1B0B8K6 A0A0K8VDK1 A0A182IAD7 A0A1A9WGC4 A0A182UYI5 A0A182SR59 A0A182XXZ3 Q9N9Y9 Q9W4T9 A0A1B0CQR5 A0A182IAD8 A0A182M1Q9 A0A182NVG0 A0A182WTP5 A0A182KDT0
Pubmed
28756777
22118469
26354079
18362917
19820115
28004739
+ More
23537049 25401762 26483478 25244985 17510324 24438588 25315136 12364791 14747013 17210077 17994087 25830018 24495485 15632085 10943839 11684659 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24485456 26109357 26109356
23537049 25401762 26483478 25244985 17510324 24438588 25315136 12364791 14747013 17210077 17994087 25830018 24495485 15632085 10943839 11684659 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24485456 26109357 26109356
EMBL
KZ149957
PZC76453.1
NWSH01000121
PCG79332.1
AGBW02010002
OWR49362.1
+ More
KQ458859 KPJ04830.1 ODYU01003674 SOQ42744.1 NWSH01003040 PCG67071.1 PCG67070.1 PCG67069.1 KPJ04832.1 KQ460254 KPJ16354.1 PZC76455.1 AGBW02010884 OWR47582.1 NWSH01000084 PCG79784.1 KQ971321 EFA00099.1 KYB28679.1 KYB28680.1 AGBW02014999 OWR40735.1 GEZM01036660 GEZM01036657 GEZM01036656 JAV82535.1 GEZM01036659 GEZM01036658 GEZM01036655 JAV82537.1 GEDC01030411 GEDC01029028 GEDC01027080 GEDC01026323 GEDC01026079 GEDC01021839 GEDC01021418 GEDC01008721 GEDC01004895 JAS06887.1 JAS08270.1 JAS10218.1 JAS10975.1 JAS11219.1 JAS15459.1 JAS15880.1 JAS28577.1 JAS32403.1 GEDC01024142 GEDC01013685 GEDC01006693 JAS13156.1 JAS23613.1 JAS30605.1 RSAL01000064 RVE49474.1 APGK01018010 APGK01018011 APGK01018012 APGK01018013 APGK01018014 APGK01018015 KB740064 ENN81761.1 KB632165 ERL89278.1 KZ150124 PZC73139.1 GFTR01007564 JAW08862.1 GBHO01024576 GBRD01006502 JAG19028.1 JAG59319.1 JXUM01018125 KQ560503 KXJ82106.1 UFQT01000015 SSX17766.1 CH477251 EAT46033.1 ATLV01013099 KE524840 KFB37203.1 AAAB01008846 EAA06357.5 CH964239 EDW81949.1 DS232044 EDS32996.1 GEBQ01005564 JAT34413.1 GBXI01005154 JAD09138.1 JXUM01051192 JXUM01051193 JXUM01051194 JXUM01051195 KQ561678 KXJ77827.1 CVRI01000013 CRK89578.1 GEBQ01016034 JAT23943.1 AXCN02002018 AXCN02002013 GAMC01011307 GAMC01011306 GAMC01011305 JAB95248.1 OUUW01000015 SPP88532.1 CH379064 EAL31716.3 KRT06480.1 JXUM01063713 KQ562264 KXJ76278.1 CCAG010019211 JXJN01009949 GDHF01015366 JAI36948.1 APCN01001155 AF196553 AJ289882 AAF86308.1 CAB96574.2 AE014298 AAF45847.2 AAN09598.1 AFH07219.1 AFH07220.1 AFH07221.1 AFH07222.1 AGB95042.1 AJWK01023927 APCN01001193 AXCM01000695
KQ458859 KPJ04830.1 ODYU01003674 SOQ42744.1 NWSH01003040 PCG67071.1 PCG67070.1 PCG67069.1 KPJ04832.1 KQ460254 KPJ16354.1 PZC76455.1 AGBW02010884 OWR47582.1 NWSH01000084 PCG79784.1 KQ971321 EFA00099.1 KYB28679.1 KYB28680.1 AGBW02014999 OWR40735.1 GEZM01036660 GEZM01036657 GEZM01036656 JAV82535.1 GEZM01036659 GEZM01036658 GEZM01036655 JAV82537.1 GEDC01030411 GEDC01029028 GEDC01027080 GEDC01026323 GEDC01026079 GEDC01021839 GEDC01021418 GEDC01008721 GEDC01004895 JAS06887.1 JAS08270.1 JAS10218.1 JAS10975.1 JAS11219.1 JAS15459.1 JAS15880.1 JAS28577.1 JAS32403.1 GEDC01024142 GEDC01013685 GEDC01006693 JAS13156.1 JAS23613.1 JAS30605.1 RSAL01000064 RVE49474.1 APGK01018010 APGK01018011 APGK01018012 APGK01018013 APGK01018014 APGK01018015 KB740064 ENN81761.1 KB632165 ERL89278.1 KZ150124 PZC73139.1 GFTR01007564 JAW08862.1 GBHO01024576 GBRD01006502 JAG19028.1 JAG59319.1 JXUM01018125 KQ560503 KXJ82106.1 UFQT01000015 SSX17766.1 CH477251 EAT46033.1 ATLV01013099 KE524840 KFB37203.1 AAAB01008846 EAA06357.5 CH964239 EDW81949.1 DS232044 EDS32996.1 GEBQ01005564 JAT34413.1 GBXI01005154 JAD09138.1 JXUM01051192 JXUM01051193 JXUM01051194 JXUM01051195 KQ561678 KXJ77827.1 CVRI01000013 CRK89578.1 GEBQ01016034 JAT23943.1 AXCN02002018 AXCN02002013 GAMC01011307 GAMC01011306 GAMC01011305 JAB95248.1 OUUW01000015 SPP88532.1 CH379064 EAL31716.3 KRT06480.1 JXUM01063713 KQ562264 KXJ76278.1 CCAG010019211 JXJN01009949 GDHF01015366 JAI36948.1 APCN01001155 AF196553 AJ289882 AAF86308.1 CAB96574.2 AE014298 AAF45847.2 AAN09598.1 AFH07219.1 AFH07220.1 AFH07221.1 AFH07222.1 AGB95042.1 AJWK01023927 APCN01001193 AXCM01000695
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000007266
UP000192223
+ More
UP000283053 UP000019118 UP000030742 UP000079169 UP000069940 UP000249989 UP000076408 UP000008820 UP000075920 UP000075885 UP000095300 UP000030765 UP000075900 UP000075880 UP000095301 UP000069272 UP000007062 UP000075903 UP000075902 UP000007798 UP000002320 UP000183832 UP000075886 UP000268350 UP000075884 UP000078200 UP000001819 UP000092445 UP000092444 UP000092443 UP000092460 UP000075840 UP000091820 UP000075901 UP000000803 UP000092461 UP000075883 UP000076407 UP000075881
UP000283053 UP000019118 UP000030742 UP000079169 UP000069940 UP000249989 UP000076408 UP000008820 UP000075920 UP000075885 UP000095300 UP000030765 UP000075900 UP000075880 UP000095301 UP000069272 UP000007062 UP000075903 UP000075902 UP000007798 UP000002320 UP000183832 UP000075886 UP000268350 UP000075884 UP000078200 UP000001819 UP000092445 UP000092444 UP000092443 UP000092460 UP000075840 UP000091820 UP000075901 UP000000803 UP000092461 UP000075883 UP000076407 UP000075881
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A2W1BXG4
A0A2A4K6N5
A0A212F6J5
A0A194QIS4
A0A2H1VR82
A0A2A4J512
+ More
A0A2A4J6J8 A0A2A4J5B3 A0A194QH44 A0A0N1IAU7 A0A2W1BS56 A0A212F1H1 A0A2A4K797 D6WHF2 A0A139WKZ8 A0A139WL53 A0A212EGX2 A0A1Y1MDY1 A0A1Y1MGV5 A0A1W4WYV5 A0A1W4WYL8 A0A1B6CCC2 A0A1B6DD38 A0A3S2LLT4 N6UM56 U4UA94 A0A2W1BD20 A0A224XJU8 A0A0A9XJP9 A0A1S4EH93 A0A182GTF8 A0A336LIT5 A0A182XXZ4 Q17HA9 A0A182W5B9 A0A182PL60 A0A1I8PJC8 A0A084VGV9 A0A182RCJ2 A0A182JDB9 A0A1I8NE46 A0A182FIZ2 Q7QEY8 A0A1I8NE48 A0A182VPE7 A0A182U923 B4NDY0 B0WQS2 A0A182J1W2 A0A1B6MEQ7 A0A182PBZ6 A0A0A1XF74 A0A182GAK6 A0A1J1HPX0 A0A1B6LJR2 A0A182RNF1 A0A182QTQ6 A0A182R0F3 A0A182W5C0 W8BE24 A0A3B0K780 A0A182NVF9 A0A1A9V0N2 Q29HP2 A0A0R3P510 A0A1A9ZLV7 A0A182GHE0 A0A1B0FNC9 A0A1A9XDY6 A0A1B0B8K6 A0A0K8VDK1 A0A182IAD7 A0A1A9WGC4 A0A182UYI5 A0A182SR59 A0A182XXZ3 Q9N9Y9 Q9W4T9 A0A1B0CQR5 A0A182IAD8 A0A182M1Q9 A0A182NVG0 A0A182WTP5 A0A182KDT0
A0A2A4J6J8 A0A2A4J5B3 A0A194QH44 A0A0N1IAU7 A0A2W1BS56 A0A212F1H1 A0A2A4K797 D6WHF2 A0A139WKZ8 A0A139WL53 A0A212EGX2 A0A1Y1MDY1 A0A1Y1MGV5 A0A1W4WYV5 A0A1W4WYL8 A0A1B6CCC2 A0A1B6DD38 A0A3S2LLT4 N6UM56 U4UA94 A0A2W1BD20 A0A224XJU8 A0A0A9XJP9 A0A1S4EH93 A0A182GTF8 A0A336LIT5 A0A182XXZ4 Q17HA9 A0A182W5B9 A0A182PL60 A0A1I8PJC8 A0A084VGV9 A0A182RCJ2 A0A182JDB9 A0A1I8NE46 A0A182FIZ2 Q7QEY8 A0A1I8NE48 A0A182VPE7 A0A182U923 B4NDY0 B0WQS2 A0A182J1W2 A0A1B6MEQ7 A0A182PBZ6 A0A0A1XF74 A0A182GAK6 A0A1J1HPX0 A0A1B6LJR2 A0A182RNF1 A0A182QTQ6 A0A182R0F3 A0A182W5C0 W8BE24 A0A3B0K780 A0A182NVF9 A0A1A9V0N2 Q29HP2 A0A0R3P510 A0A1A9ZLV7 A0A182GHE0 A0A1B0FNC9 A0A1A9XDY6 A0A1B0B8K6 A0A0K8VDK1 A0A182IAD7 A0A1A9WGC4 A0A182UYI5 A0A182SR59 A0A182XXZ3 Q9N9Y9 Q9W4T9 A0A1B0CQR5 A0A182IAD8 A0A182M1Q9 A0A182NVG0 A0A182WTP5 A0A182KDT0
PDB
4OF8
E-value=1.0144e-84,
Score=802
Ontologies
GO
Topology
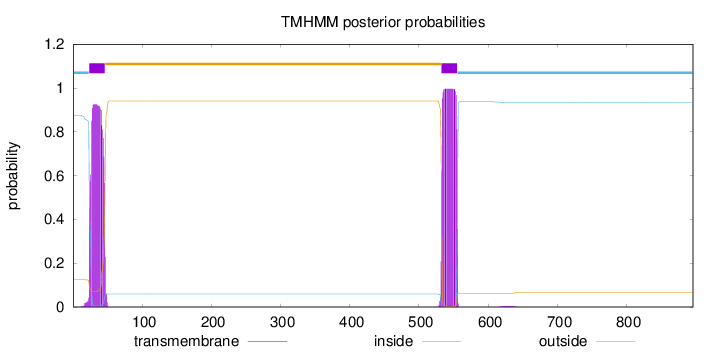
Length:
896
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
42.5674800000002
Exp number, first 60 AAs:
19.56903
Total prob of N-in:
0.87598
POSSIBLE N-term signal
sequence
inside
1 - 23
TMhelix
24 - 46
outside
47 - 532
TMhelix
533 - 555
inside
556 - 896
Population Genetic Test Statistics
Pi
240.588641
Theta
172.340197
Tajima's D
1.197832
CLR
0.139761
CSRT
0.71986400679966
Interpretation
Uncertain
