Pre Gene Modal
BGIBMGA013297
Annotation
PREDICTED:_ryanodine_receptor_44F_isoform_X23_[Bombyx_mori]
Full name
Ryanodine receptor
Alternative Name
Ryanodine receptor 44F
Location in the cell
Cytoplasmic Reliability : 2.255
Sequence
CDS
ATGGTGTGCCTCTCGTGTACCGCTACCGGGGAACGCGTGTGCCTCGCAGCTGAAGGATTTGGAAACCGGCACTGCTTCCTGGAGAACATCGCGGACAAGAACATACCACCCGATCTGTCGCAATGCGTCTTCGTCATAGAACAGGCGTTATCAGTGAGAGCTCTCCAAGAATTGGTGACGGCGGCCGGATCGGAAACCGGCAAGGGCACTGGATCCGGTCACAGGACACTGCTTTACGGAAACGCGATCCTTTTGAGACATCTCAACAGTGATATGTACTTGGCGTGCTTGTCGACATCATCATCTCAAGACAAACTGGCTTTCGACGTTGGTCTCCAAGAGCACTCCCAGGGTGAAGCGTGCTGGTGGACATTGCATCCGGCCAGTAAACAAAGATCGGAGGGTGAAAAAGTGCGAGTGGGAGATGATTTGATTCTTGTATCTGTTGCCACTGAGAGATATTTGCACACAACTAAAGAGAATGAAGTATCGATCGTCAATGCCTCGTTTCACGTGACACACTGGTCGGTCCAGCCATACGGAACCGGCATCTCGAGGATGAAATACGTCGGCTACGTCTTCGGGGGAGACGTTCTACGTTTCTTCCACGGAGGAGACGAATGCCTTACAATACCTAGCACTTGGACTCGGGATGGAGGACAGAATATTGTAGTATACGAAGGCGGATCGGTCATGTCACAAGCTCGTTCGCTATGGCGTTTGGAACTAGCTCGTACCAAATGGGCCGGAGGCTTTATCAACTGGTACCATCCAATGAGGATTCGGCACATCACTACGGGTCGATACTTGGGTGTTAACGACCAGAATGAACTCCATTTGGTCAGCAGAGAGGAAGCCACAACCGCTTCCTGCGCCTTTTGTCTTCGTCAAGAAAAAGATGATCAGAAACAAGTGCTTGAAGATAAAGATTTGGAAGTGATCGGAGCGCCAATTATTAAATACGGTGACTCAACAGTAATTGTGCAGCACTCTGAAACTGGCTTGTGGCTTTCATATAAGTCTTACGAAACAAAGAAGAAAGGCGTCGGTAAAGTAGAAGAGAAACAAGCCATACTACACGAAGAAGGTAAAATGGACGACGGCCTTGATTTCTCCAGGTCCCAAGAAGAAGAGTCGAGGACCGCCAGAGTCATCAGGAAGTGTTCTTCTTTATTCACTAAGTTCATCAATGGCCTTGAAACTCTTCAAGAAAACAGAAGACACTCGATGTTCTTCGCGTCCGTGAACTTAGGAGAGATGGTGATGTGCTTGGAGGATCTTATAAACTATTTCGCACAGCCCGATGAGGACATGGAACACGAAGAAAAGCAAAACAAGTTCCGTGCTCTTCGAAACCGCCAAGATCTTTTCCAAGAGGAGGGTATCCTGAATTTAATCCTGGAAGCAATCGACAAGATCAACGTGATCACGTCCCAGGGATTCCTTGCTGGGTTCCTAGCTGGAGACGAATCGGGACAGAGCTGGGAAATGATCTCTGGATACCTTTATCAGCTTTTAGCTGCTATTATAAAAGGAAACCACACGAACTGCGCACAGTTCGCAAACTCCAACCGTCTGAACTGGTTGTTCTCTAGACTCGGCTCACAGGCATCTGGCGAAGGAACTGGGATGCTGGACGTGTTGCATTGCGTGCTCATAGACTCGCCTGAAGCCTTGAATATGATGAGGGATGAGCACATAAAAGTAATCATAAGCTTATTAGAAAAGCACGGGCGTGACCCGAAAGTTCTCGACGTGCTGTGCTCGCTGTGCGTCGGCAACGGAGTGGCCGTGCGCTCGTCCCAGAACAACATCTGCGACTACTTGCTGCCAGGGAAGAACTTGCTACTGCAGACTCAGCTAGTAGACCACGTTTCCAGTGTGCGACCGAACATTTTCGTGGGCCGCGTCGAAGGATCAGCAGTGTACCGTAAATGGTACTTCGAAGTGACGATGGACCACATCGAGAAAACCACGCACATGATGCCCCATCTGCGCATAGGATGGGCTAACACTTCTGGATACGTACCGTATCCTGGTGGTGGCGAAAAGTGGGGCGGTAACGGTGTCGGTGATGATTTGTATTCCTTTGGTTTCGATGGAGCATATTTGTGGTCTGGTGGCAGGAAAACCCCGGTTAATCGCACTCACCACGAAGAACCTTATATCAGAAAAGGCGATGTGATTGGGTGCGCTCTGGACTTGAACGTTCCTTTAATCAACTTCATGTTCAACGGCGTCAGGGTGACCGGATCTTTCACCAACTTCAACTTGGAAGGCATGTTCTTCCCCGCCATCAGCTGCTCAAGTAAATTAAGTTGTCGGTTTCTATTCGGCGGCGAACATGGTCGTCTGCGTTACGCGGCTCCCGAGGGATACTCTCCGCTGGTTGAGTCGCTGCTTCCGCAACAGATCCTCAGTCTCGAACCCTGCTTCTATTTTGGCAACCTCAACAAGAGGGCTCTAGCTGGACCATCTTTAGTTCAAGATGATACCGCATTTGTGCCTACACCGGTTGATACGATGACGATATCTCTGCCAACATACGTGGAACAAATCAGGGATAAGCTAGCCGAAAATATTCACGAAATGTGGGCTATGAATAAGATTGAAGCCGGCTGGATGTACGGTGATCAGCGCGACGACATACATAAGATCCATCCTTGCTTGGTGCCCTTCGAGCGACTACCACCTGCTGAGAAGAGATACGATATCCAACTAGCTGTGCAAACATTGAAAACAATTCTTGCGCTTGGCTACTACATTACTTTGGATAAGCCACCAGCACGTATCCGCAACGTCCGACTTCCAAACGAACCATTTTTACAGTCAAACGGCTATAAACCAGCACCTTTAGACCTTGGTGCTGTTACTCTGACACCCAAAATGGATGAACTAGTGGACCAATTAGCTGAGAATACTCACAATCTTTGGGCCAGGGAAAGAATACAACAAGGTTGGACTTACGGTCTCAATGAGGATCCGGAGATGCATCGCTCCCCTCACTTAGTGCCGTATCCTAAAGTTGATGATGCCATAAAAAAAGCCAACCGAGACACCGCGTCTGAAACTGTCCGAACTTTATTGGTTTACGGATATAATCTGGATCCGCCCACTGGAGAACAGCATGAAGCTCTACTAGCGGAGGCTTCTAAGCAGAAACAAGCTGATTTCAGAACCTACAGGGCGGAAAAGAACTACGCCGTTAGTTCTGGGAAATGGTATTTCGAATTTGAAATATTAACGGCTGGGCCAATGAGGGTTGGTTGGGCTCATGCTGACATGGCTCCGGGCATGATGTTGGGACAAGACGAGAATTCTTGGGCGTTCGACGGATACAACTGCTTAAAATTACATGGCGGAACGAGCGACGCATTCGGTATACAGCTCAAAGTCGGGGATATTGTGGGCTGCTTCCTAGACGTCGTCGACCAAACAATTAGTGAGTCGATGGCTTAA
Protein
MVCLSCTATGERVCLAAEGFGNRHCFLENIADKNIPPDLSQCVFVIEQALSVRALQELVTAAGSETGKGTGSGHRTLLYGNAILLRHLNSDMYLACLSTSSSQDKLAFDVGLQEHSQGEACWWTLHPASKQRSEGEKVRVGDDLILVSVATERYLHTTKENEVSIVNASFHVTHWSVQPYGTGISRMKYVGYVFGGDVLRFFHGGDECLTIPSTWTRDGGQNIVVYEGGSVMSQARSLWRLELARTKWAGGFINWYHPMRIRHITTGRYLGVNDQNELHLVSREEATTASCAFCLRQEKDDQKQVLEDKDLEVIGAPIIKYGDSTVIVQHSETGLWLSYKSYETKKKGVGKVEEKQAILHEEGKMDDGLDFSRSQEEESRTARVIRKCSSLFTKFINGLETLQENRRHSMFFASVNLGEMVMCLEDLINYFAQPDEDMEHEEKQNKFRALRNRQDLFQEEGILNLILEAIDKINVITSQGFLAGFLAGDESGQSWEMISGYLYQLLAAIIKGNHTNCAQFANSNRLNWLFSRLGSQASGEGTGMLDVLHCVLIDSPEALNMMRDEHIKVIISLLEKHGRDPKVLDVLCSLCVGNGVAVRSSQNNICDYLLPGKNLLLQTQLVDHVSSVRPNIFVGRVEGSAVYRKWYFEVTMDHIEKTTHMMPHLRIGWANTSGYVPYPGGGEKWGGNGVGDDLYSFGFDGAYLWSGGRKTPVNRTHHEEPYIRKGDVIGCALDLNVPLINFMFNGVRVTGSFTNFNLEGMFFPAISCSSKLSCRFLFGGEHGRLRYAAPEGYSPLVESLLPQQILSLEPCFYFGNLNKRALAGPSLVQDDTAFVPTPVDTMTISLPTYVEQIRDKLAENIHEMWAMNKIEAGWMYGDQRDDIHKIHPCLVPFERLPPAEKRYDIQLAVQTLKTILALGYYITLDKPPARIRNVRLPNEPFLQSNGYKPAPLDLGAVTLTPKMDELVDQLAENTHNLWARERIQQGWTYGLNEDPEMHRSPHLVPYPKVDDAIKKANRDTASETVRTLLVYGYNLDPPTGEQHEALLAEASKQKQADFRTYRAEKNYAVSSGKWYFEFEILTAGPMRVGWAHADMAPGMMLGQDENSWAFDGYNCLKLHGGTSDAFGIQLKVGDIVGCFLDVVDQTISESMA
Summary
Description
Intracellular calcium channel that is required for proper muscle function during embryonic development and may be essential for excitation-contraction coupling in larval body wall muscles.
Subunit
Homotetramer.
Miscellaneous
Channel activity is modulated by the alkaloid ryanodine that binds to the open Ca-release channel with high affinity and maintains the channel in an open conformation. The calcium release channel is modulated by calcium ions, magnesium ions, ATP and calmodulin (By similarity).
Similarity
Belongs to the ryanodine receptor (TC 1.A.3.1) family.
Keywords
Alternative splicing
Calcium
Calcium channel
Calcium transport
Complete proteome
Developmental protein
Ion channel
Ion transport
Ligand-gated ion channel
Membrane
Phosphoprotein
Receptor
Reference proteome
Repeat
Sarcoplasmic reticulum
Transmembrane
Transmembrane helix
Transport
Feature
chain Ryanodine receptor
splice variant In isoform C and isoform D.
splice variant In isoform C and isoform D.
Uniprot
I3VR31
H9CNL0
A0A0P0QM12
A0A2Z4EVR6
A0A0P0QLR0
A0A0P0QLT4
+ More
V5RE97 A0A2A4JFR2 A0A212F8V6 A0A194QMY4 A0A0N7JQQ1 I6ZY07 M4T4G3 A0A2Z5BST2 A0A0P0QM17 A0A164LU87 I1XB02 A0A059XRP6 A0A059XMJ0 I3VR34 A0A059XHF1 A0A0E3XBL1 A0A2H4QGA9 A0A0H4P5L6 A0A0P0QM39 A0A0P0QM29 A0A1J0I9Q3 A0A0P0QLU2 A0A0S1Z233 A0A2H4QG92 A0A0P0QM31 X2GG79 A0A089FYX0 A0A0P0QLS4 G8IJI2 K7W8M2 G8EME4 G8EME3 I3NWV8 I3VR32 R9R5D5 A0A2A4JFT6 A0A097P7A6 A0A139WKL1 D6WF72 A0A158P0A3 A0A088AV96 A0A2A3ESZ8 A0A151IVU0 E2BZJ9 A0A195FCN9 A0A151K2J5 A0A195B655 A0A3L8D605 A0A059XRQ0 U3PWV7 A0A059XRL5 A0A023NE85 I3VR35 A0A059XLU6 A0A1I8MDA4 A0A1I8MDA1 A0A1I8MD94 A0A1I8MD98 A0A1I8MDB0 A0A026W7Q8 A0A1I8MD97 A0A0L0BUZ9 A0A1I8P9D0 A0A1I8P9K7 A0A1I8P9E0 A0A1I8P9D4 A0A1I8P9D5 A0A1I8MDA2 A0A1I8MDA3 A0A1I8MD99 A0A1I8MDA9 A0A1I8MD93 A0A023NEE5 A0A0Q9WXJ7 A0A1I8MDA6 B4MK53 A0A1I8MDB1 A0A0Q9X8Q5 A0A0Q9XKI2 A0A0R1DUL1 A0A0Q9XIC2 A0A0Q9X8Q4 A0A0R1DMA2 A0A0R1DTC6 A0A0R1DTS7 A0A0Q9X8P8 A0A0B4K715 A0A0R1DM53 B4P3C9 Q24498 Q24498-3 A0A0B4K7K0 A0A0B4K7U9 A0A1W4UJL1
V5RE97 A0A2A4JFR2 A0A212F8V6 A0A194QMY4 A0A0N7JQQ1 I6ZY07 M4T4G3 A0A2Z5BST2 A0A0P0QM17 A0A164LU87 I1XB02 A0A059XRP6 A0A059XMJ0 I3VR34 A0A059XHF1 A0A0E3XBL1 A0A2H4QGA9 A0A0H4P5L6 A0A0P0QM39 A0A0P0QM29 A0A1J0I9Q3 A0A0P0QLU2 A0A0S1Z233 A0A2H4QG92 A0A0P0QM31 X2GG79 A0A089FYX0 A0A0P0QLS4 G8IJI2 K7W8M2 G8EME4 G8EME3 I3NWV8 I3VR32 R9R5D5 A0A2A4JFT6 A0A097P7A6 A0A139WKL1 D6WF72 A0A158P0A3 A0A088AV96 A0A2A3ESZ8 A0A151IVU0 E2BZJ9 A0A195FCN9 A0A151K2J5 A0A195B655 A0A3L8D605 A0A059XRQ0 U3PWV7 A0A059XRL5 A0A023NE85 I3VR35 A0A059XLU6 A0A1I8MDA4 A0A1I8MDA1 A0A1I8MD94 A0A1I8MD98 A0A1I8MDB0 A0A026W7Q8 A0A1I8MD97 A0A0L0BUZ9 A0A1I8P9D0 A0A1I8P9K7 A0A1I8P9E0 A0A1I8P9D4 A0A1I8P9D5 A0A1I8MDA2 A0A1I8MDA3 A0A1I8MD99 A0A1I8MDA9 A0A1I8MD93 A0A023NEE5 A0A0Q9WXJ7 A0A1I8MDA6 B4MK53 A0A1I8MDB1 A0A0Q9X8Q5 A0A0Q9XKI2 A0A0R1DUL1 A0A0Q9XIC2 A0A0Q9X8Q4 A0A0R1DMA2 A0A0R1DTC6 A0A0R1DTS7 A0A0Q9X8P8 A0A0B4K715 A0A0R1DM53 B4P3C9 Q24498 Q24498-3 A0A0B4K7K0 A0A0B4K7U9 A0A1W4UJL1
Pubmed
29891375
24267694
22118469
26354079
23603125
24098400
+ More
29626986 22567170 27845250 22982600 22761165 29191465 23010195 25330781 18362917 19820115 21347285 20798317 30249741 24607641 25315136 24508170 26108605 24485316 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 8276118 1338312 10811919 18327897
29626986 22567170 27845250 22982600 22761165 29191465 23010195 25330781 18362917 19820115 21347285 20798317 30249741 24607641 25315136 24508170 26108605 24485316 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 8276118 1338312 10811919 18327897
EMBL
JQ769302
AFK84955.1
JQ354988
AFC36359.1
KR815505
ALL55471.1
+ More
MG712298 AWV67093.1 KR815501 ALL55467.1 KR815506 ALL55472.1 KF641862 AHB33498.1 NWSH01001744 PCG70263.1 AGBW02009672 OWR50158.1 KQ458859 KPJ04841.1 KR815502 ALL55468.1 JX082287 AFN70719.1 KC355370 AGH68757.1 MF623806 AXA98483.1 KR815499 ALL55465.1 KR088972 AMN82804.1 JQ799046 AFI80904.1 KJ573633 AIA23854.1 KJ573634 AIA23855.1 JQ769305 AFK84958.1 KJ573635 AIA23856.1 KP213290 AKC03558.2 MG013971 ATX64238.1 KP721560 AKP49172.1 KR815507 ALL55473.1 KR815503 ALL55469.1 KX519762 APC65631.1 KR815504 ALL55470.1 KT029915 ALM96708.1 MG013970 ATX64237.1 KR815498 ALL55464.1 KJ135037 AHN16453.1 KM034750 AIP90097.1 KR815500 ALL55466.1 JN801028 AET09964.1 JX467684 AFW97408.1 JF926694 AER25355.1 JF926693 AER25354.1 JF927788 AEI91094.1 JQ769303 AFK84956.1 JX481776 AGI62938.1 PCG70262.1 KM216386 AIU40166.1 KQ971327 KYB28433.1 EEZ99829.2 ADTU01004756 ADTU01004757 KZ288192 PBC34336.1 KQ980881 KYN11971.1 GL451627 EFN78897.1 KQ981685 KYN37967.1 LKEX01025115 KYN50349.1 KQ976587 KYM79740.1 QOIP01000012 RLU15601.1 KJ573638 AIA23859.1 KF306296 AGW82429.1 KJ573637 AIA23858.1 KF734670 AHW99830.1 JQ769306 AFK84959.1 KJ573636 AIA23857.1 KK107353 EZA52107.1 JRES01001421 KNC23059.1 KF734669 AHW99829.1 CH963846 KRF97582.1 EDW72492.2 CH933808 KRG04800.1 KRG04791.1 CM000157 KRJ98422.1 KRG04786.1 KRG04790.1 KRJ98394.1 KRJ98400.1 KRJ98403.1 KRG04795.1 AE013599 AFH07961.1 KRJ98399.1 EDW89402.1 D17389 Z18536 AAF59036.2 AFH07963.1 AFH07964.1
MG712298 AWV67093.1 KR815501 ALL55467.1 KR815506 ALL55472.1 KF641862 AHB33498.1 NWSH01001744 PCG70263.1 AGBW02009672 OWR50158.1 KQ458859 KPJ04841.1 KR815502 ALL55468.1 JX082287 AFN70719.1 KC355370 AGH68757.1 MF623806 AXA98483.1 KR815499 ALL55465.1 KR088972 AMN82804.1 JQ799046 AFI80904.1 KJ573633 AIA23854.1 KJ573634 AIA23855.1 JQ769305 AFK84958.1 KJ573635 AIA23856.1 KP213290 AKC03558.2 MG013971 ATX64238.1 KP721560 AKP49172.1 KR815507 ALL55473.1 KR815503 ALL55469.1 KX519762 APC65631.1 KR815504 ALL55470.1 KT029915 ALM96708.1 MG013970 ATX64237.1 KR815498 ALL55464.1 KJ135037 AHN16453.1 KM034750 AIP90097.1 KR815500 ALL55466.1 JN801028 AET09964.1 JX467684 AFW97408.1 JF926694 AER25355.1 JF926693 AER25354.1 JF927788 AEI91094.1 JQ769303 AFK84956.1 JX481776 AGI62938.1 PCG70262.1 KM216386 AIU40166.1 KQ971327 KYB28433.1 EEZ99829.2 ADTU01004756 ADTU01004757 KZ288192 PBC34336.1 KQ980881 KYN11971.1 GL451627 EFN78897.1 KQ981685 KYN37967.1 LKEX01025115 KYN50349.1 KQ976587 KYM79740.1 QOIP01000012 RLU15601.1 KJ573638 AIA23859.1 KF306296 AGW82429.1 KJ573637 AIA23858.1 KF734670 AHW99830.1 JQ769306 AFK84959.1 KJ573636 AIA23857.1 KK107353 EZA52107.1 JRES01001421 KNC23059.1 KF734669 AHW99829.1 CH963846 KRF97582.1 EDW72492.2 CH933808 KRG04800.1 KRG04791.1 CM000157 KRJ98422.1 KRG04786.1 KRG04790.1 KRJ98394.1 KRJ98400.1 KRJ98403.1 KRG04795.1 AE013599 AFH07961.1 KRJ98399.1 EDW89402.1 D17389 Z18536 AAF59036.2 AFH07963.1 AFH07964.1
Proteomes
PRIDE
Pfam
Interpro
IPR003877
SPRY_dom
+ More
IPR013333 Ryan_recept
IPR003032 Ryanodine_rcpt
IPR016093 MIR_motif
IPR035761 SPRY1_RyR
IPR036300 MIR_dom_sf
IPR001870 B30.2/SPRY
IPR013662 RIH_assoc-dom
IPR000699 RIH_dom
IPR014821 Ins145_P3_rcpt
IPR011992 EF-hand-dom_pair
IPR035762 SPRY3_RyR
IPR013320 ConA-like_dom_sf
IPR035910 RyR/IP3R_RIH_dom_sf
IPR009460 Ryanrecept_TM4-6
IPR035764 SPRY2_RyR
IPR005821 Ion_trans_dom
IPR018247 EF_Hand_1_Ca_BS
IPR002048 EF_hand_dom
IPR016024 ARM-type_fold
IPR013333 Ryan_recept
IPR003032 Ryanodine_rcpt
IPR016093 MIR_motif
IPR035761 SPRY1_RyR
IPR036300 MIR_dom_sf
IPR001870 B30.2/SPRY
IPR013662 RIH_assoc-dom
IPR000699 RIH_dom
IPR014821 Ins145_P3_rcpt
IPR011992 EF-hand-dom_pair
IPR035762 SPRY3_RyR
IPR013320 ConA-like_dom_sf
IPR035910 RyR/IP3R_RIH_dom_sf
IPR009460 Ryanrecept_TM4-6
IPR035764 SPRY2_RyR
IPR005821 Ion_trans_dom
IPR018247 EF_Hand_1_Ca_BS
IPR002048 EF_hand_dom
IPR016024 ARM-type_fold
SUPFAM
ProteinModelPortal
I3VR31
H9CNL0
A0A0P0QM12
A0A2Z4EVR6
A0A0P0QLR0
A0A0P0QLT4
+ More
V5RE97 A0A2A4JFR2 A0A212F8V6 A0A194QMY4 A0A0N7JQQ1 I6ZY07 M4T4G3 A0A2Z5BST2 A0A0P0QM17 A0A164LU87 I1XB02 A0A059XRP6 A0A059XMJ0 I3VR34 A0A059XHF1 A0A0E3XBL1 A0A2H4QGA9 A0A0H4P5L6 A0A0P0QM39 A0A0P0QM29 A0A1J0I9Q3 A0A0P0QLU2 A0A0S1Z233 A0A2H4QG92 A0A0P0QM31 X2GG79 A0A089FYX0 A0A0P0QLS4 G8IJI2 K7W8M2 G8EME4 G8EME3 I3NWV8 I3VR32 R9R5D5 A0A2A4JFT6 A0A097P7A6 A0A139WKL1 D6WF72 A0A158P0A3 A0A088AV96 A0A2A3ESZ8 A0A151IVU0 E2BZJ9 A0A195FCN9 A0A151K2J5 A0A195B655 A0A3L8D605 A0A059XRQ0 U3PWV7 A0A059XRL5 A0A023NE85 I3VR35 A0A059XLU6 A0A1I8MDA4 A0A1I8MDA1 A0A1I8MD94 A0A1I8MD98 A0A1I8MDB0 A0A026W7Q8 A0A1I8MD97 A0A0L0BUZ9 A0A1I8P9D0 A0A1I8P9K7 A0A1I8P9E0 A0A1I8P9D4 A0A1I8P9D5 A0A1I8MDA2 A0A1I8MDA3 A0A1I8MD99 A0A1I8MDA9 A0A1I8MD93 A0A023NEE5 A0A0Q9WXJ7 A0A1I8MDA6 B4MK53 A0A1I8MDB1 A0A0Q9X8Q5 A0A0Q9XKI2 A0A0R1DUL1 A0A0Q9XIC2 A0A0Q9X8Q4 A0A0R1DMA2 A0A0R1DTC6 A0A0R1DTS7 A0A0Q9X8P8 A0A0B4K715 A0A0R1DM53 B4P3C9 Q24498 Q24498-3 A0A0B4K7K0 A0A0B4K7U9 A0A1W4UJL1
V5RE97 A0A2A4JFR2 A0A212F8V6 A0A194QMY4 A0A0N7JQQ1 I6ZY07 M4T4G3 A0A2Z5BST2 A0A0P0QM17 A0A164LU87 I1XB02 A0A059XRP6 A0A059XMJ0 I3VR34 A0A059XHF1 A0A0E3XBL1 A0A2H4QGA9 A0A0H4P5L6 A0A0P0QM39 A0A0P0QM29 A0A1J0I9Q3 A0A0P0QLU2 A0A0S1Z233 A0A2H4QG92 A0A0P0QM31 X2GG79 A0A089FYX0 A0A0P0QLS4 G8IJI2 K7W8M2 G8EME4 G8EME3 I3NWV8 I3VR32 R9R5D5 A0A2A4JFT6 A0A097P7A6 A0A139WKL1 D6WF72 A0A158P0A3 A0A088AV96 A0A2A3ESZ8 A0A151IVU0 E2BZJ9 A0A195FCN9 A0A151K2J5 A0A195B655 A0A3L8D605 A0A059XRQ0 U3PWV7 A0A059XRL5 A0A023NE85 I3VR35 A0A059XLU6 A0A1I8MDA4 A0A1I8MDA1 A0A1I8MD94 A0A1I8MD98 A0A1I8MDB0 A0A026W7Q8 A0A1I8MD97 A0A0L0BUZ9 A0A1I8P9D0 A0A1I8P9K7 A0A1I8P9E0 A0A1I8P9D4 A0A1I8P9D5 A0A1I8MDA2 A0A1I8MDA3 A0A1I8MD99 A0A1I8MDA9 A0A1I8MD93 A0A023NEE5 A0A0Q9WXJ7 A0A1I8MDA6 B4MK53 A0A1I8MDB1 A0A0Q9X8Q5 A0A0Q9XKI2 A0A0R1DUL1 A0A0Q9XIC2 A0A0Q9X8Q4 A0A0R1DMA2 A0A0R1DTC6 A0A0R1DTS7 A0A0Q9X8P8 A0A0B4K715 A0A0R1DM53 B4P3C9 Q24498 Q24498-3 A0A0B4K7K0 A0A0B4K7U9 A0A1W4UJL1
PDB
5GOA
E-value=0,
Score=3099
Ontologies
GO
Topology
Subcellular location
Sarcoplasmic reticulum membrane
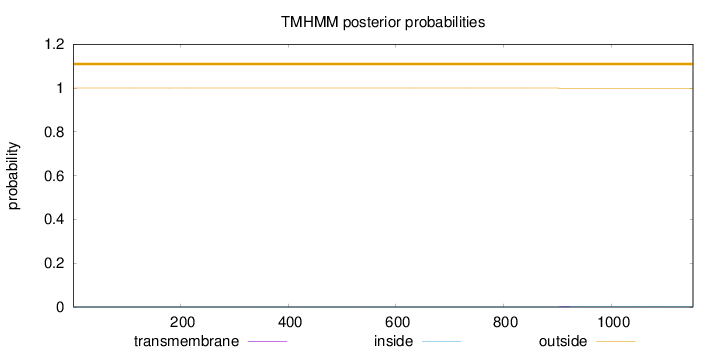
The number of predicted transmembrane domains varies between orthologs, but both N-terminus and C-terminus seem to be cytoplasmic. With evidence from 1 publications.
Membrane The number of predicted transmembrane domains varies between orthologs, but both N-terminus and C-terminus seem to be cytoplasmic. With evidence from 1 publications.
Membrane The number of predicted transmembrane domains varies between orthologs, but both N-terminus and C-terminus seem to be cytoplasmic. With evidence from 1 publications.
Length:
1152
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03627
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.00020
outside
1 - 1152
Population Genetic Test Statistics
Pi
212.910035
Theta
20.093291
Tajima's D
0.181077
CLR
1.152531
CSRT
0.429728513574321
Interpretation
Uncertain
