Gene
KWMTBOMO14366
Pre Gene Modal
BGIBMGA013305
Annotation
PREDICTED:_Golgi_pH_regulator_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.801
Sequence
CDS
ATGTCTTTCTTAGAAGATACGTTAATAATTTTAGGATCTCAGGTAGTATTCTTCGTTGGAGGGTGGATTTTCTTCGTAAAACAACTGTTCCGTGATTATGAAGTACATCATACACTCGTACAGCTTATATTCTCTGTTACGTTCGCCCTAAGCTGTACTATGTTCGAGCTGATCATATTCGAAATCATTGGCTATTTAGATACAAGTTCCCGTTACTTTCATTGGAATTTAGGCTTATATTCACTGCTGTTTATGGTGATAGCTCTCATACCCTTCTACATTGCTTTTTTCTGCATAAGCAACATTAGATTTGTGTCACATAACTCCATTAGGCCTCTCACCGTGTTTGTATGGTTCATATACTTGTATTTCTTTTGGAAAATCGGCGATCCATTTCCAATACTGAGTCCTAAAAAGGGCATACTCTCGATAGAGCAAGGAGTTTCAAGGATCGGTGTCATCGGTGTGACTGTCATGGCTCTTCTGTCAGGTTTCGGTGCTGTCAACTATCCTTACACTTCAATGGCTATATTTATAAGACCAGTAACCCAATCCGATGTACTATCAATAGAAAAGAAACTGCTCCAAACAATGGACATGATATTGGTTAAGAAGAAGAGGATAGCTTTAGCTGAAGCGAACAGCGTTGGTGGGAGACAGTATAATATGTTGGACCAATCTGGAACGAAAAATAGAGGTTTCTGGACAAACATTCTGTCCAGCGTTGGAAGTATTGCGAATCCCTTAGGTCATGGAACGGAAAATATTAGTCAATTGAGGCAAGAGATATCCGGCTTAGAGGAGCTGAGCCGTCAATTGTTCTTAGAAGCGCACGACGCCAGAACGATGAGAGAAAAAATCGAATGGTCGAAGACTTTTCAAGGGAAATATTTCAATTTCTTAGGTTATTTCTTCTCTTTGTATTGTATATGGAAGATATTTATTTCCACCGTGAACATAGTATTCGACAGAGTCGGCAAAAAGGATCCGGTCACAAAAGGCCTAGAGATAGTCGTCCACTGGCTCGGTTGGAACATAGATGTCGCTTTTTGGTCTCAGCACGTCTCTTTCATACTCGTCGGCTGTATAGTGCTCACCAGCATCAGAGGTTTGCTGCTGACGTTGACGAAGTTCTTCTACAAGATCTCATCATCGAAATCATCGAATATAATCGTTTTAATACTGGCCCAGATAATGGGAATGTATTTCTGTTCGTCAGTGCTGCTGATGAGAATGAACATGCCCCCCGAATACCGCATCATAATAACTCAAGTCCTCGGCGAATTACAGTTCAACTTCTATCACAGATGGTTCGACGTAATATTTTTAGTTAGCGCACTGACCAGTATATTTACACTGTATTTAGCCCACAAACAGCCGTCGGTCAATTAA
Protein
MSFLEDTLIILGSQVVFFVGGWIFFVKQLFRDYEVHHTLVQLIFSVTFALSCTMFELIIFEIIGYLDTSSRYFHWNLGLYSLLFMVIALIPFYIAFFCISNIRFVSHNSIRPLTVFVWFIYLYFFWKIGDPFPILSPKKGILSIEQGVSRIGVIGVTVMALLSGFGAVNYPYTSMAIFIRPVTQSDVLSIEKKLLQTMDMILVKKKRIALAEANSVGGRQYNMLDQSGTKNRGFWTNILSSVGSIANPLGHGTENISQLRQEISGLEELSRQLFLEAHDARTMREKIEWSKTFQGKYFNFLGYFFSLYCIWKIFISTVNIVFDRVGKKDPVTKGLEIVVHWLGWNIDVAFWSQHVSFILVGCIVLTSIRGLLLTLTKFFYKISSSKSSNIIVLILAQIMGMYFCSSVLLMRMNMPPEYRIIITQVLGELQFNFYHRWFDVIFLVSALTSIFTLYLAHKQPSVN
Summary
Uniprot
A0A194QHE8
A0A2A4K2I7
A0A3S2P0R6
A0A194RIG3
H9JUU2
A0A067QTY0
+ More
A0A2J7PH25 A0A1B6KGU9 S4PE41 A0A158NDK8 A0A212ERA8 A0A088A809 K7J9B6 A0A0M8ZZD9 A0A1B6GW51 A0A195BP51 A0A195DB96 A0A151XEH0 A0A069DUJ1 A0A195F3K6 E2A0H9 A0A195CMZ9 A0A023F4Z3 A0A3L8DZU1 E9IU83 A0A0P4VM54 T1IDN0 A0A232FLY0 A0A1E1W5I3 E0VJL9 A0A0A9XC65 B0XHU3 A0A1Q3F6R3 D2CFW7 A0A2R5LI15 Q16KQ4 A0A1W4WJS1 A0A182WR70 A0A182FUX6 A0A1S4H771 A0A1Y9GLP0 A0A182VDH9 A0A1L8DK11 U5EKS7 A0A182TEY4 A0A084VEV9 A0A182RXN4 A0A2M4AIC4 A0A2M4BMW7 A0A182MEG0 A0A293N453 A0A182SQY1 A0A182P346 A0A0P5FTA0 W5JWQ9 A0A182QB45 A0A131YM02 A0A182YE80 L7M8M7 A0A1E1X3S1 A0A023GNC4 G3MPD1 T1JAQ6 A0A182WDA7 A0A224YZ18 A0A182K4S8 A0A0P4ZF48 Q7PSQ7 A0A131XEE9 A0A0L7RB93 A0A023FMF7 F4W548 A0A1D2NML6 E2BII7 A0A0N8A330 A0A131XZ81 A0A023FXD6 A0A0P5FBK7 A0A0K8R4B3 A0A0P5G7S5 A0A026WIM3 A0A154PN25 A0A182N342 A0A1B6CG01 A0A1Y1MWL2 A0A3Q2WDH9 I3KAD0 A0A3P8NN47 A0A3P9AVI4 A0A3Q4GIX7 A0A3P9MTR0 A0A1W4ZXM1 A0A3B4Y437 A0A3B5B9G1 A0A3B3DB00 A0A3B4TKE7 A0A3Q1EHR8 A0A3B3V3F1 A0A1S2X113 A0A3P8RKI2
A0A2J7PH25 A0A1B6KGU9 S4PE41 A0A158NDK8 A0A212ERA8 A0A088A809 K7J9B6 A0A0M8ZZD9 A0A1B6GW51 A0A195BP51 A0A195DB96 A0A151XEH0 A0A069DUJ1 A0A195F3K6 E2A0H9 A0A195CMZ9 A0A023F4Z3 A0A3L8DZU1 E9IU83 A0A0P4VM54 T1IDN0 A0A232FLY0 A0A1E1W5I3 E0VJL9 A0A0A9XC65 B0XHU3 A0A1Q3F6R3 D2CFW7 A0A2R5LI15 Q16KQ4 A0A1W4WJS1 A0A182WR70 A0A182FUX6 A0A1S4H771 A0A1Y9GLP0 A0A182VDH9 A0A1L8DK11 U5EKS7 A0A182TEY4 A0A084VEV9 A0A182RXN4 A0A2M4AIC4 A0A2M4BMW7 A0A182MEG0 A0A293N453 A0A182SQY1 A0A182P346 A0A0P5FTA0 W5JWQ9 A0A182QB45 A0A131YM02 A0A182YE80 L7M8M7 A0A1E1X3S1 A0A023GNC4 G3MPD1 T1JAQ6 A0A182WDA7 A0A224YZ18 A0A182K4S8 A0A0P4ZF48 Q7PSQ7 A0A131XEE9 A0A0L7RB93 A0A023FMF7 F4W548 A0A1D2NML6 E2BII7 A0A0N8A330 A0A131XZ81 A0A023FXD6 A0A0P5FBK7 A0A0K8R4B3 A0A0P5G7S5 A0A026WIM3 A0A154PN25 A0A182N342 A0A1B6CG01 A0A1Y1MWL2 A0A3Q2WDH9 I3KAD0 A0A3P8NN47 A0A3P9AVI4 A0A3Q4GIX7 A0A3P9MTR0 A0A1W4ZXM1 A0A3B4Y437 A0A3B5B9G1 A0A3B3DB00 A0A3B4TKE7 A0A3Q1EHR8 A0A3B3V3F1 A0A1S2X113 A0A3P8RKI2
Pubmed
26354079
19121390
24845553
23622113
21347285
22118469
+ More
20075255 26334808 20798317 25474469 30249741 21282665 27129103 28648823 20566863 25401762 18362917 19820115 17510324 12364791 24438588 20920257 23761445 26830274 25244985 25576852 28503490 22216098 28797301 28049606 21719571 27289101 24508170 28004739 25186727 29451363 20433749
20075255 26334808 20798317 25474469 30249741 21282665 27129103 28648823 20566863 25401762 18362917 19820115 17510324 12364791 24438588 20920257 23761445 26830274 25244985 25576852 28503490 22216098 28797301 28049606 21719571 27289101 24508170 28004739 25186727 29451363 20433749
EMBL
KQ458859
KPJ04844.1
NWSH01000255
PCG77983.1
RSAL01000064
RVE49451.1
+ More
KQ460124 KPJ17613.1 BABH01031037 KK852954 KDR13284.1 NEVH01025150 PNF15632.1 GEBQ01029304 JAT10673.1 GAIX01004712 JAA87848.1 ADTU01012699 AGBW02013061 OWR44033.1 KQ435793 KOX74220.1 GECZ01003123 JAS66646.1 KQ976424 KYM88290.1 KQ981082 KYN09689.1 KQ982254 KYQ58761.1 GBGD01001418 JAC87471.1 KQ981820 KYN35165.1 GL435626 EFN72882.1 KQ977600 KYN01464.1 GBBI01002628 JAC16084.1 QOIP01000002 RLU25328.1 GL765847 EFZ15867.1 GDKW01000990 JAI55605.1 ACPB03012772 NNAY01000034 OXU31751.1 GDQN01008816 JAT82238.1 DS235226 EEB13575.1 GBHO01028949 GBRD01017075 GBRD01014993 JAG14655.1 JAG48752.1 DS233208 EDS28666.1 GFDL01011803 JAV23242.1 DS497670 EFA13037.1 GGLE01005036 MBY09162.1 CH477943 EAT34887.1 AAAB01008816 APCN01005462 GFDF01007323 JAV06761.1 GANO01001693 JAB58178.1 ATLV01012338 KE524783 KFB36503.1 GGFK01007218 MBW40539.1 GGFJ01005221 MBW54362.1 AXCM01006844 GFWV01023177 MAA47904.1 GDIQ01251750 GDIP01132387 LRGB01001005 JAJ99974.1 JAL71327.1 KZS14093.1 ADMH02000001 ETN68158.1 AXCN02001130 GEDV01008570 JAP79987.1 GACK01004822 JAA60212.1 GFAC01005268 JAT93920.1 GBBM01001053 JAC34365.1 JO843732 AEO35349.1 JH432002 GFPF01007864 MAA19010.1 GDIP01213576 JAJ09826.1 EAA05189.5 GEFH01004133 JAP64448.1 KQ414617 KOC68139.1 GBBK01002452 JAC22030.1 GL887596 EGI70744.1 LJIJ01000005 ODN06477.1 GL448512 EFN84497.1 GDIP01187297 JAJ36105.1 GEFM01003253 JAP72543.1 GBBL01001128 JAC26192.1 GDIQ01256768 JAJ94956.1 GADI01007912 JAA65896.1 GDIQ01245359 JAK06366.1 KK107200 EZA55506.1 KQ434977 KZC12864.1 GEDC01024995 JAS12303.1 GEZM01021551 JAV88950.1 AERX01029986
KQ460124 KPJ17613.1 BABH01031037 KK852954 KDR13284.1 NEVH01025150 PNF15632.1 GEBQ01029304 JAT10673.1 GAIX01004712 JAA87848.1 ADTU01012699 AGBW02013061 OWR44033.1 KQ435793 KOX74220.1 GECZ01003123 JAS66646.1 KQ976424 KYM88290.1 KQ981082 KYN09689.1 KQ982254 KYQ58761.1 GBGD01001418 JAC87471.1 KQ981820 KYN35165.1 GL435626 EFN72882.1 KQ977600 KYN01464.1 GBBI01002628 JAC16084.1 QOIP01000002 RLU25328.1 GL765847 EFZ15867.1 GDKW01000990 JAI55605.1 ACPB03012772 NNAY01000034 OXU31751.1 GDQN01008816 JAT82238.1 DS235226 EEB13575.1 GBHO01028949 GBRD01017075 GBRD01014993 JAG14655.1 JAG48752.1 DS233208 EDS28666.1 GFDL01011803 JAV23242.1 DS497670 EFA13037.1 GGLE01005036 MBY09162.1 CH477943 EAT34887.1 AAAB01008816 APCN01005462 GFDF01007323 JAV06761.1 GANO01001693 JAB58178.1 ATLV01012338 KE524783 KFB36503.1 GGFK01007218 MBW40539.1 GGFJ01005221 MBW54362.1 AXCM01006844 GFWV01023177 MAA47904.1 GDIQ01251750 GDIP01132387 LRGB01001005 JAJ99974.1 JAL71327.1 KZS14093.1 ADMH02000001 ETN68158.1 AXCN02001130 GEDV01008570 JAP79987.1 GACK01004822 JAA60212.1 GFAC01005268 JAT93920.1 GBBM01001053 JAC34365.1 JO843732 AEO35349.1 JH432002 GFPF01007864 MAA19010.1 GDIP01213576 JAJ09826.1 EAA05189.5 GEFH01004133 JAP64448.1 KQ414617 KOC68139.1 GBBK01002452 JAC22030.1 GL887596 EGI70744.1 LJIJ01000005 ODN06477.1 GL448512 EFN84497.1 GDIP01187297 JAJ36105.1 GEFM01003253 JAP72543.1 GBBL01001128 JAC26192.1 GDIQ01256768 JAJ94956.1 GADI01007912 JAA65896.1 GDIQ01245359 JAK06366.1 KK107200 EZA55506.1 KQ434977 KZC12864.1 GEDC01024995 JAS12303.1 GEZM01021551 JAV88950.1 AERX01029986
Proteomes
UP000053268
UP000218220
UP000283053
UP000053240
UP000005204
UP000027135
+ More
UP000235965 UP000005205 UP000007151 UP000005203 UP000002358 UP000053105 UP000078540 UP000078492 UP000075809 UP000078541 UP000000311 UP000078542 UP000279307 UP000015103 UP000215335 UP000009046 UP000002320 UP000007266 UP000008820 UP000192223 UP000076407 UP000069272 UP000075840 UP000075903 UP000075902 UP000030765 UP000075900 UP000075883 UP000075901 UP000075885 UP000076858 UP000000673 UP000075886 UP000076408 UP000075920 UP000075881 UP000007062 UP000053825 UP000007755 UP000094527 UP000008237 UP000053097 UP000076502 UP000075884 UP000264840 UP000005207 UP000265100 UP000265160 UP000261580 UP000242638 UP000192224 UP000261360 UP000261400 UP000261560 UP000261420 UP000257200 UP000261500 UP000087266 UP000265080
UP000235965 UP000005205 UP000007151 UP000005203 UP000002358 UP000053105 UP000078540 UP000078492 UP000075809 UP000078541 UP000000311 UP000078542 UP000279307 UP000015103 UP000215335 UP000009046 UP000002320 UP000007266 UP000008820 UP000192223 UP000076407 UP000069272 UP000075840 UP000075903 UP000075902 UP000030765 UP000075900 UP000075883 UP000075901 UP000075885 UP000076858 UP000000673 UP000075886 UP000076408 UP000075920 UP000075881 UP000007062 UP000053825 UP000007755 UP000094527 UP000008237 UP000053097 UP000076502 UP000075884 UP000264840 UP000005207 UP000265100 UP000265160 UP000261580 UP000242638 UP000192224 UP000261360 UP000261400 UP000261560 UP000261420 UP000257200 UP000261500 UP000087266 UP000265080
PRIDE
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
A0A194QHE8
A0A2A4K2I7
A0A3S2P0R6
A0A194RIG3
H9JUU2
A0A067QTY0
+ More
A0A2J7PH25 A0A1B6KGU9 S4PE41 A0A158NDK8 A0A212ERA8 A0A088A809 K7J9B6 A0A0M8ZZD9 A0A1B6GW51 A0A195BP51 A0A195DB96 A0A151XEH0 A0A069DUJ1 A0A195F3K6 E2A0H9 A0A195CMZ9 A0A023F4Z3 A0A3L8DZU1 E9IU83 A0A0P4VM54 T1IDN0 A0A232FLY0 A0A1E1W5I3 E0VJL9 A0A0A9XC65 B0XHU3 A0A1Q3F6R3 D2CFW7 A0A2R5LI15 Q16KQ4 A0A1W4WJS1 A0A182WR70 A0A182FUX6 A0A1S4H771 A0A1Y9GLP0 A0A182VDH9 A0A1L8DK11 U5EKS7 A0A182TEY4 A0A084VEV9 A0A182RXN4 A0A2M4AIC4 A0A2M4BMW7 A0A182MEG0 A0A293N453 A0A182SQY1 A0A182P346 A0A0P5FTA0 W5JWQ9 A0A182QB45 A0A131YM02 A0A182YE80 L7M8M7 A0A1E1X3S1 A0A023GNC4 G3MPD1 T1JAQ6 A0A182WDA7 A0A224YZ18 A0A182K4S8 A0A0P4ZF48 Q7PSQ7 A0A131XEE9 A0A0L7RB93 A0A023FMF7 F4W548 A0A1D2NML6 E2BII7 A0A0N8A330 A0A131XZ81 A0A023FXD6 A0A0P5FBK7 A0A0K8R4B3 A0A0P5G7S5 A0A026WIM3 A0A154PN25 A0A182N342 A0A1B6CG01 A0A1Y1MWL2 A0A3Q2WDH9 I3KAD0 A0A3P8NN47 A0A3P9AVI4 A0A3Q4GIX7 A0A3P9MTR0 A0A1W4ZXM1 A0A3B4Y437 A0A3B5B9G1 A0A3B3DB00 A0A3B4TKE7 A0A3Q1EHR8 A0A3B3V3F1 A0A1S2X113 A0A3P8RKI2
A0A2J7PH25 A0A1B6KGU9 S4PE41 A0A158NDK8 A0A212ERA8 A0A088A809 K7J9B6 A0A0M8ZZD9 A0A1B6GW51 A0A195BP51 A0A195DB96 A0A151XEH0 A0A069DUJ1 A0A195F3K6 E2A0H9 A0A195CMZ9 A0A023F4Z3 A0A3L8DZU1 E9IU83 A0A0P4VM54 T1IDN0 A0A232FLY0 A0A1E1W5I3 E0VJL9 A0A0A9XC65 B0XHU3 A0A1Q3F6R3 D2CFW7 A0A2R5LI15 Q16KQ4 A0A1W4WJS1 A0A182WR70 A0A182FUX6 A0A1S4H771 A0A1Y9GLP0 A0A182VDH9 A0A1L8DK11 U5EKS7 A0A182TEY4 A0A084VEV9 A0A182RXN4 A0A2M4AIC4 A0A2M4BMW7 A0A182MEG0 A0A293N453 A0A182SQY1 A0A182P346 A0A0P5FTA0 W5JWQ9 A0A182QB45 A0A131YM02 A0A182YE80 L7M8M7 A0A1E1X3S1 A0A023GNC4 G3MPD1 T1JAQ6 A0A182WDA7 A0A224YZ18 A0A182K4S8 A0A0P4ZF48 Q7PSQ7 A0A131XEE9 A0A0L7RB93 A0A023FMF7 F4W548 A0A1D2NML6 E2BII7 A0A0N8A330 A0A131XZ81 A0A023FXD6 A0A0P5FBK7 A0A0K8R4B3 A0A0P5G7S5 A0A026WIM3 A0A154PN25 A0A182N342 A0A1B6CG01 A0A1Y1MWL2 A0A3Q2WDH9 I3KAD0 A0A3P8NN47 A0A3P9AVI4 A0A3Q4GIX7 A0A3P9MTR0 A0A1W4ZXM1 A0A3B4Y437 A0A3B5B9G1 A0A3B3DB00 A0A3B4TKE7 A0A3Q1EHR8 A0A3B3V3F1 A0A1S2X113 A0A3P8RKI2
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
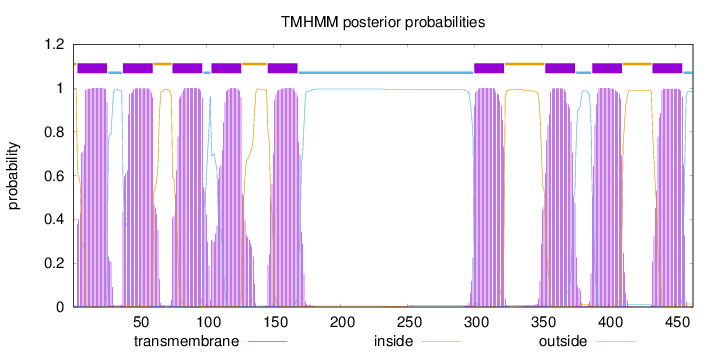
Length:
463
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
198.12879
Exp number, first 60 AAs:
42.00436
Total prob of N-in:
0.00692
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 26
inside
27 - 37
TMhelix
38 - 60
outside
61 - 74
TMhelix
75 - 97
inside
98 - 103
TMhelix
104 - 126
outside
127 - 145
TMhelix
146 - 168
inside
169 - 299
TMhelix
300 - 322
outside
323 - 352
TMhelix
353 - 375
inside
376 - 387
TMhelix
388 - 410
outside
411 - 432
TMhelix
433 - 455
inside
456 - 463
Population Genetic Test Statistics
Pi
297.781717
Theta
194.554326
Tajima's D
1.997455
CLR
0.177569
CSRT
0.880005999700015
Interpretation
Uncertain
