Pre Gene Modal
BGIBMGA013299
Annotation
glycerol_kinase-like_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.584
Sequence
CDS
ATGCCAGTCCGGGACGGCTCCAAGAAGCCCCTGGTCGCTGTTATAGATGACGGAACCAAGACCGTCAGATTTGTAATATACGAAGCGGAATGCTCGGAGGAACTGGCCTCGTATCAGATGGACAAGACGGAGGTGCAGCCCCACGAGGGTTGGTCAGAGCAAGATCCATACGAGATAATGCACCACATAAAGCTTTGTGCTGAAAATGCCATTGACCAGTTAACAGAATTAGGATATTCAAAAGATGACATCATTACTTTAGGCATAACGAATCAGAGGGAGACAACTATAGCTTGGGACAAATACACCGGGGAGCCATTGCACCCCGCCATAGCCTGGAACGACATACGTACCGATAGCACGGTAGACGCGATCCTAGCCAAAGTGCCGGACCGGAATAAGAATTATCTGAAGAACATCTGCGGTCTACCGATATCGCCTTACTTCAGCGCGCTGAAGATGCGCTGGTTGAAGGACAACGTGAAGGCAGTCAGACGAGCGAGTAGAGATAAGCGATTGCTATTCGGCACTGTGGACTCGTGGATTCTGTGGAATTTAACCGGCGGTCCCCAGGACGGTCTTCACGTGACCGACGTGACGAACGCTTCGAGGACTTTGCTCATGAATCTGGAGACCCTGAATTGGGACCCCGTGCTGTGTCGGCTATTCGGAATCCATCGGGATACGTTGGCGGAGATAAGAAGTAGCTCCGAAGTGTACGGCACGGTCAGAGACGGCTCCGTTCTCGACGGCATATCCATTTCAGGGATTTTAGGTAACCAGCAATCTGCGCTCGTGGGTCAGAACTGCTTGAGCGCCGGCCAAGCGAAGAACACTTACCGCAGCGGGTGCTTTCTGCTGTACAACACCGGAACCAGACGGGTCAATTCGACCCACGGTCTGATCACGACGATAGCTTATAAATTAGGACCCGATCAACCCCCCATTTACGCATTGGAGGGTTCGATCGCGGTGGCGGGCGGCGCTATGAAGTTCCTAAGGGACAACCTGAGGCTGATAAGGGACGTGTCTCAGGACACTGAGCACATCGCGGGCCAGGTGTTCTCCACCGGGGACGTGTACTTCGTGCCGGCCTTCAGTGGCCTCTACGCGCCCTATTGGAGGAAGGACGCCAGAGGTATAATCTGCGGCCTGACCGCCTTCACGACGAAGAACCACATAATACGGGCCGGATTGGAAGCGGTGTGCTTTCAGACTAGGGACATCCTGGAAGCGATGAACAAGGACTGCGGGATGCCCCTGTCGAAGCTCCACGTGGACGGGAAGATGTCCTCGAACGATCTGCTGATGCAGCTACAGGCGGACCTCACCGGTATACCGGTTTTGCGCGCCCAGTCGTGGGACATGTCCGCTCTGGGCGTGGGCATCGTGGCGGGCCACTCGGTGGGCGTGTGGAGCTGCGACAAGTGGAAGCATCACGCCACCCACGCCGACACCTTCCTGCCGACCACGACCGATGATGATCGCGATGCCAGATACACGAAATGGAAGATGGCCGTCCAGCGCTCCCTCGGATGGGCGACCACGAAGAAGTCGATCACAATGACAGAGGAAAGGTATAAGCTCCTATCGTCGATACCCGCAGCTTTGTACCTCATAGGGAGCTTTACGATGCTAGTCGCCTCGTCGATTTTAAACGAAGCACGTAGTTGA
Protein
MPVRDGSKKPLVAVIDDGTKTVRFVIYEAECSEELASYQMDKTEVQPHEGWSEQDPYEIMHHIKLCAENAIDQLTELGYSKDDIITLGITNQRETTIAWDKYTGEPLHPAIAWNDIRTDSTVDAILAKVPDRNKNYLKNICGLPISPYFSALKMRWLKDNVKAVRRASRDKRLLFGTVDSWILWNLTGGPQDGLHVTDVTNASRTLLMNLETLNWDPVLCRLFGIHRDTLAEIRSSSEVYGTVRDGSVLDGISISGILGNQQSALVGQNCLSAGQAKNTYRSGCFLLYNTGTRRVNSTHGLITTIAYKLGPDQPPIYALEGSIAVAGGAMKFLRDNLRLIRDVSQDTEHIAGQVFSTGDVYFVPAFSGLYAPYWRKDARGIICGLTAFTTKNHIIRAGLEAVCFQTRDILEAMNKDCGMPLSKLHVDGKMSSNDLLMQLQADLTGIPVLRAQSWDMSALGVGIVAGHSVGVWSCDKWKHHATHADTFLPTTTDDDRDARYTKWKMAVQRSLGWATTKKSITMTEERYKLLSSIPAALYLIGSFTMLVASSILNEARS
Summary
Similarity
Belongs to the FGGY kinase family.
Uniprot
B0I1G6
A0A2A4K133
A0A2H1W351
A0A2D3E1J6
A0A2A4K1N9
A0A2A4K2S5
+ More
A0A194Q6K2 A0A0N1IFZ9 H9JUT6 A0A2W1B823 D6WF03 A0A1S4FCX1 Q176X4 A0A1L8E224 A0A088A7T5 A0A1Y1N5U1 A0A026W800 A0A0C9QQE1 A0A0C9PWP6 A0A158NIJ5 E2A7C0 A0A182VWK6 A0A182WSD2 A0A182UTU4 A0A182IX57 A0A0M9A124 A0A182L9L6 A0A2C9GQX5 Q8T5I3 A0A182YAD5 A0A182R4T5 A0A1B6EXR0 A0A2A3ENU1 J3JXE0 A0A1Q3G2D7 A0A195CJI8 A0A0L7RB57 A0A067QXW3 A0A084VAH3 A0A0K8T8F0 A0A154PNI9 A0A0A9XQH6 A0A0K8T8D8 A0A2M4BJD0 E2C8G9 E9I8E6 W5JJS5 A0A182Q2C4 A0A151K0Z3 A0A1B6L7W5 A0A151XFW3 A0A023F717 K7J5W5 A0A182PGT0 A0A182LY18 A0A1B6DPI0 A0A182FQD3 A0A151JSM7 A0A182N5V1 A0A232FF01 A0A0K8T8D7 A0A0K8T9H3 A0A0A9XTH1 A0A1B6D684 A0A195BCV5 A0A224XAQ2 A0A0P4VMT7 T1I3E8 A0A069DZ73 A0A0V0G4N7 T1PG73 A0A1I8M0Y8 A0A1I8M0Z1 A0A0L0CCC3 A0A1I8P3H7 A0A1I8P3B7 E0VR80 A0A0M3QWJ2 B4KXV6 B4MGN3 A0A0R3P969 A0A212EN04 A0A0R3P6J6 B4IYM8 A0A2J7R7K1 A0A3B0JXW0 A0A3B0JW06 B4MMQ6 Q29E10 B4H433 A0A0R3P6G4 F4WPZ8 A0A1W4VVG5 A0A0J9RLZ2 Q961K9 Q9W095 B4HVZ4 B4PDT2 A0A067RJF6 A0A1A9ZFV2
A0A194Q6K2 A0A0N1IFZ9 H9JUT6 A0A2W1B823 D6WF03 A0A1S4FCX1 Q176X4 A0A1L8E224 A0A088A7T5 A0A1Y1N5U1 A0A026W800 A0A0C9QQE1 A0A0C9PWP6 A0A158NIJ5 E2A7C0 A0A182VWK6 A0A182WSD2 A0A182UTU4 A0A182IX57 A0A0M9A124 A0A182L9L6 A0A2C9GQX5 Q8T5I3 A0A182YAD5 A0A182R4T5 A0A1B6EXR0 A0A2A3ENU1 J3JXE0 A0A1Q3G2D7 A0A195CJI8 A0A0L7RB57 A0A067QXW3 A0A084VAH3 A0A0K8T8F0 A0A154PNI9 A0A0A9XQH6 A0A0K8T8D8 A0A2M4BJD0 E2C8G9 E9I8E6 W5JJS5 A0A182Q2C4 A0A151K0Z3 A0A1B6L7W5 A0A151XFW3 A0A023F717 K7J5W5 A0A182PGT0 A0A182LY18 A0A1B6DPI0 A0A182FQD3 A0A151JSM7 A0A182N5V1 A0A232FF01 A0A0K8T8D7 A0A0K8T9H3 A0A0A9XTH1 A0A1B6D684 A0A195BCV5 A0A224XAQ2 A0A0P4VMT7 T1I3E8 A0A069DZ73 A0A0V0G4N7 T1PG73 A0A1I8M0Y8 A0A1I8M0Z1 A0A0L0CCC3 A0A1I8P3H7 A0A1I8P3B7 E0VR80 A0A0M3QWJ2 B4KXV6 B4MGN3 A0A0R3P969 A0A212EN04 A0A0R3P6J6 B4IYM8 A0A2J7R7K1 A0A3B0JXW0 A0A3B0JW06 B4MMQ6 Q29E10 B4H433 A0A0R3P6G4 F4WPZ8 A0A1W4VVG5 A0A0J9RLZ2 Q961K9 Q9W095 B4HVZ4 B4PDT2 A0A067RJF6 A0A1A9ZFV2
Pubmed
19748584
26354079
19121390
28756777
18362917
19820115
+ More
17510324 28004739 24508170 30249741 21347285 20798317 20966253 12060762 12364791 14747013 17210077 25244985 22516182 23537049 24845553 24438588 25401762 26823975 21282665 20920257 23761445 25474469 20075255 28648823 27129103 26334808 25315136 26108605 20566863 17994087 18057021 15632085 22118469 23185243 21719571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
17510324 28004739 24508170 30249741 21347285 20798317 20966253 12060762 12364791 14747013 17210077 25244985 22516182 23537049 24845553 24438588 25401762 26823975 21282665 20920257 23761445 25474469 20075255 28648823 27129103 26334808 25315136 26108605 20566863 17994087 18057021 15632085 22118469 23185243 21719571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
AB250689
BAG06923.2
NWSH01000255
PCG77981.1
ODYU01006038
SOQ47525.1
+ More
KY565575 ATU31387.1 PCG77979.1 PCG77980.1 KQ459386 KPJ01167.1 KQ460601 KPJ13810.1 BABH01031032 BABH01031033 BABH01031034 KZ150233 PZC71989.1 KQ971319 EFA00308.1 CH477381 EAT42198.1 GFDF01001378 JAV12706.1 GEZM01012077 GEZM01012075 JAV93231.1 KK107347 QOIP01000007 EZA52232.1 RLU20105.1 GBYB01005924 GBYB01005932 JAG75691.1 JAG75699.1 GBYB01005928 JAG75695.1 ADTU01016861 GL437329 EFN70617.1 KQ435793 KOX74206.1 APCN01000883 AJ439353 AAAB01008987 CAD27929.1 EAA00896.1 GECZ01027000 JAS42769.1 KZ288203 PBC33387.1 APGK01050051 APGK01050052 APGK01050053 APGK01050054 BT127909 KB741165 AEE62871.1 ENN73409.1 GFDL01001104 JAV33941.1 KQ977642 KYN00901.1 KQ414617 KOC68152.1 KK852846 KDR15118.1 ATLV01003737 KE524159 KFB34967.1 GBRD01004012 GBRD01004010 JAG61809.1 KQ434977 KZC12878.1 GBHO01020617 JAG22987.1 GBRD01009028 GBRD01004008 GDHC01020711 GDHC01016810 GDHC01003879 JAG61813.1 JAP97917.1 JAQ01819.1 JAQ14750.1 GGFJ01003920 MBW53061.1 GL453666 EFN75699.1 GL761538 EFZ23161.1 ADMH02001287 ETN63155.1 AXCN02001151 KQ981298 KYN43066.1 GEBQ01031240 GEBQ01030941 GEBQ01030284 GEBQ01030192 GEBQ01023912 GEBQ01020170 GEBQ01015303 GEBQ01015250 GEBQ01014826 GEBQ01014512 GEBQ01013449 GEBQ01012473 GEBQ01010210 GEBQ01007055 GEBQ01003223 GEBQ01002301 JAT08737.1 JAT09036.1 JAT09693.1 JAT09785.1 JAT16065.1 JAT19807.1 JAT24674.1 JAT24727.1 JAT25151.1 JAT25465.1 JAT26528.1 JAT27504.1 JAT29767.1 JAT32922.1 JAT36754.1 JAT37676.1 KQ982179 KYQ59279.1 GBBI01001718 JAC16994.1 AXCM01006311 GEDC01009731 GEDC01003636 JAS27567.1 JAS33662.1 KQ978557 KYN30151.1 NNAY01000305 OXU29334.1 GBRD01004009 JAG61812.1 GBRD01004011 JAG61810.1 GBHO01020618 JAG22986.1 GEDC01030161 GEDC01016109 JAS07137.1 JAS21189.1 KQ976513 KYM82401.1 GFTR01006960 JAW09466.1 GDKW01002957 JAI53638.1 ACPB03010438 GBGD01001140 JAC87749.1 GECL01003205 JAP02919.1 KA647812 AFP62441.1 JRES01000611 KNC29881.1 DS235459 EEB15886.1 CP012525 ALC44243.1 CH933809 EDW17628.1 KRG05652.1 KRG05653.1 KRG05654.1 KRG05655.1 KRG05656.1 CH940682 EDW57099.1 KRF77577.1 KRF77578.1 KRF77579.1 KRF77580.1 CH379070 KRT08752.1 AGBW02013754 OWR42874.1 KRT08753.1 CH916366 EDV95538.1 NEVH01006731 PNF36814.1 OUUW01000009 SPP85262.1 SPP85263.1 CH963847 EDW73462.1 EAL30253.1 KRT08751.1 CH479208 EDW31148.1 KRT08754.1 GL888261 EGI63727.1 CM002912 KMY96933.1 KMY96934.1 KMY96935.1 KMY96936.1 AY051535 AAK92959.1 AE014296 BT044165 AAF47558.2 AAF47559.1 AAF47560.1 AAN11496.1 AAN11497.1 ACH92230.1 AHN57929.1 CH480817 EDW50109.1 CM000159 EDW92897.1 KRK00789.1 KK852595 KDR20628.1
KY565575 ATU31387.1 PCG77979.1 PCG77980.1 KQ459386 KPJ01167.1 KQ460601 KPJ13810.1 BABH01031032 BABH01031033 BABH01031034 KZ150233 PZC71989.1 KQ971319 EFA00308.1 CH477381 EAT42198.1 GFDF01001378 JAV12706.1 GEZM01012077 GEZM01012075 JAV93231.1 KK107347 QOIP01000007 EZA52232.1 RLU20105.1 GBYB01005924 GBYB01005932 JAG75691.1 JAG75699.1 GBYB01005928 JAG75695.1 ADTU01016861 GL437329 EFN70617.1 KQ435793 KOX74206.1 APCN01000883 AJ439353 AAAB01008987 CAD27929.1 EAA00896.1 GECZ01027000 JAS42769.1 KZ288203 PBC33387.1 APGK01050051 APGK01050052 APGK01050053 APGK01050054 BT127909 KB741165 AEE62871.1 ENN73409.1 GFDL01001104 JAV33941.1 KQ977642 KYN00901.1 KQ414617 KOC68152.1 KK852846 KDR15118.1 ATLV01003737 KE524159 KFB34967.1 GBRD01004012 GBRD01004010 JAG61809.1 KQ434977 KZC12878.1 GBHO01020617 JAG22987.1 GBRD01009028 GBRD01004008 GDHC01020711 GDHC01016810 GDHC01003879 JAG61813.1 JAP97917.1 JAQ01819.1 JAQ14750.1 GGFJ01003920 MBW53061.1 GL453666 EFN75699.1 GL761538 EFZ23161.1 ADMH02001287 ETN63155.1 AXCN02001151 KQ981298 KYN43066.1 GEBQ01031240 GEBQ01030941 GEBQ01030284 GEBQ01030192 GEBQ01023912 GEBQ01020170 GEBQ01015303 GEBQ01015250 GEBQ01014826 GEBQ01014512 GEBQ01013449 GEBQ01012473 GEBQ01010210 GEBQ01007055 GEBQ01003223 GEBQ01002301 JAT08737.1 JAT09036.1 JAT09693.1 JAT09785.1 JAT16065.1 JAT19807.1 JAT24674.1 JAT24727.1 JAT25151.1 JAT25465.1 JAT26528.1 JAT27504.1 JAT29767.1 JAT32922.1 JAT36754.1 JAT37676.1 KQ982179 KYQ59279.1 GBBI01001718 JAC16994.1 AXCM01006311 GEDC01009731 GEDC01003636 JAS27567.1 JAS33662.1 KQ978557 KYN30151.1 NNAY01000305 OXU29334.1 GBRD01004009 JAG61812.1 GBRD01004011 JAG61810.1 GBHO01020618 JAG22986.1 GEDC01030161 GEDC01016109 JAS07137.1 JAS21189.1 KQ976513 KYM82401.1 GFTR01006960 JAW09466.1 GDKW01002957 JAI53638.1 ACPB03010438 GBGD01001140 JAC87749.1 GECL01003205 JAP02919.1 KA647812 AFP62441.1 JRES01000611 KNC29881.1 DS235459 EEB15886.1 CP012525 ALC44243.1 CH933809 EDW17628.1 KRG05652.1 KRG05653.1 KRG05654.1 KRG05655.1 KRG05656.1 CH940682 EDW57099.1 KRF77577.1 KRF77578.1 KRF77579.1 KRF77580.1 CH379070 KRT08752.1 AGBW02013754 OWR42874.1 KRT08753.1 CH916366 EDV95538.1 NEVH01006731 PNF36814.1 OUUW01000009 SPP85262.1 SPP85263.1 CH963847 EDW73462.1 EAL30253.1 KRT08751.1 CH479208 EDW31148.1 KRT08754.1 GL888261 EGI63727.1 CM002912 KMY96933.1 KMY96934.1 KMY96935.1 KMY96936.1 AY051535 AAK92959.1 AE014296 BT044165 AAF47558.2 AAF47559.1 AAF47560.1 AAN11496.1 AAN11497.1 ACH92230.1 AHN57929.1 CH480817 EDW50109.1 CM000159 EDW92897.1 KRK00789.1 KK852595 KDR20628.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000005204
UP000007266
UP000008820
+ More
UP000005203 UP000053097 UP000279307 UP000005205 UP000000311 UP000075920 UP000076407 UP000075903 UP000075880 UP000053105 UP000075882 UP000075840 UP000007062 UP000076408 UP000075900 UP000242457 UP000019118 UP000078542 UP000053825 UP000027135 UP000030765 UP000076502 UP000008237 UP000000673 UP000075886 UP000078541 UP000075809 UP000002358 UP000075885 UP000075883 UP000069272 UP000078492 UP000075884 UP000215335 UP000078540 UP000015103 UP000095301 UP000037069 UP000095300 UP000009046 UP000092553 UP000009192 UP000008792 UP000001819 UP000007151 UP000001070 UP000235965 UP000268350 UP000007798 UP000008744 UP000007755 UP000192221 UP000000803 UP000001292 UP000002282 UP000092445
UP000005203 UP000053097 UP000279307 UP000005205 UP000000311 UP000075920 UP000076407 UP000075903 UP000075880 UP000053105 UP000075882 UP000075840 UP000007062 UP000076408 UP000075900 UP000242457 UP000019118 UP000078542 UP000053825 UP000027135 UP000030765 UP000076502 UP000008237 UP000000673 UP000075886 UP000078541 UP000075809 UP000002358 UP000075885 UP000075883 UP000069272 UP000078492 UP000075884 UP000215335 UP000078540 UP000015103 UP000095301 UP000037069 UP000095300 UP000009046 UP000092553 UP000009192 UP000008792 UP000001819 UP000007151 UP000001070 UP000235965 UP000268350 UP000007798 UP000008744 UP000007755 UP000192221 UP000000803 UP000001292 UP000002282 UP000092445
Interpro
ProteinModelPortal
B0I1G6
A0A2A4K133
A0A2H1W351
A0A2D3E1J6
A0A2A4K1N9
A0A2A4K2S5
+ More
A0A194Q6K2 A0A0N1IFZ9 H9JUT6 A0A2W1B823 D6WF03 A0A1S4FCX1 Q176X4 A0A1L8E224 A0A088A7T5 A0A1Y1N5U1 A0A026W800 A0A0C9QQE1 A0A0C9PWP6 A0A158NIJ5 E2A7C0 A0A182VWK6 A0A182WSD2 A0A182UTU4 A0A182IX57 A0A0M9A124 A0A182L9L6 A0A2C9GQX5 Q8T5I3 A0A182YAD5 A0A182R4T5 A0A1B6EXR0 A0A2A3ENU1 J3JXE0 A0A1Q3G2D7 A0A195CJI8 A0A0L7RB57 A0A067QXW3 A0A084VAH3 A0A0K8T8F0 A0A154PNI9 A0A0A9XQH6 A0A0K8T8D8 A0A2M4BJD0 E2C8G9 E9I8E6 W5JJS5 A0A182Q2C4 A0A151K0Z3 A0A1B6L7W5 A0A151XFW3 A0A023F717 K7J5W5 A0A182PGT0 A0A182LY18 A0A1B6DPI0 A0A182FQD3 A0A151JSM7 A0A182N5V1 A0A232FF01 A0A0K8T8D7 A0A0K8T9H3 A0A0A9XTH1 A0A1B6D684 A0A195BCV5 A0A224XAQ2 A0A0P4VMT7 T1I3E8 A0A069DZ73 A0A0V0G4N7 T1PG73 A0A1I8M0Y8 A0A1I8M0Z1 A0A0L0CCC3 A0A1I8P3H7 A0A1I8P3B7 E0VR80 A0A0M3QWJ2 B4KXV6 B4MGN3 A0A0R3P969 A0A212EN04 A0A0R3P6J6 B4IYM8 A0A2J7R7K1 A0A3B0JXW0 A0A3B0JW06 B4MMQ6 Q29E10 B4H433 A0A0R3P6G4 F4WPZ8 A0A1W4VVG5 A0A0J9RLZ2 Q961K9 Q9W095 B4HVZ4 B4PDT2 A0A067RJF6 A0A1A9ZFV2
A0A194Q6K2 A0A0N1IFZ9 H9JUT6 A0A2W1B823 D6WF03 A0A1S4FCX1 Q176X4 A0A1L8E224 A0A088A7T5 A0A1Y1N5U1 A0A026W800 A0A0C9QQE1 A0A0C9PWP6 A0A158NIJ5 E2A7C0 A0A182VWK6 A0A182WSD2 A0A182UTU4 A0A182IX57 A0A0M9A124 A0A182L9L6 A0A2C9GQX5 Q8T5I3 A0A182YAD5 A0A182R4T5 A0A1B6EXR0 A0A2A3ENU1 J3JXE0 A0A1Q3G2D7 A0A195CJI8 A0A0L7RB57 A0A067QXW3 A0A084VAH3 A0A0K8T8F0 A0A154PNI9 A0A0A9XQH6 A0A0K8T8D8 A0A2M4BJD0 E2C8G9 E9I8E6 W5JJS5 A0A182Q2C4 A0A151K0Z3 A0A1B6L7W5 A0A151XFW3 A0A023F717 K7J5W5 A0A182PGT0 A0A182LY18 A0A1B6DPI0 A0A182FQD3 A0A151JSM7 A0A182N5V1 A0A232FF01 A0A0K8T8D7 A0A0K8T9H3 A0A0A9XTH1 A0A1B6D684 A0A195BCV5 A0A224XAQ2 A0A0P4VMT7 T1I3E8 A0A069DZ73 A0A0V0G4N7 T1PG73 A0A1I8M0Y8 A0A1I8M0Z1 A0A0L0CCC3 A0A1I8P3H7 A0A1I8P3B7 E0VR80 A0A0M3QWJ2 B4KXV6 B4MGN3 A0A0R3P969 A0A212EN04 A0A0R3P6J6 B4IYM8 A0A2J7R7K1 A0A3B0JXW0 A0A3B0JW06 B4MMQ6 Q29E10 B4H433 A0A0R3P6G4 F4WPZ8 A0A1W4VVG5 A0A0J9RLZ2 Q961K9 Q9W095 B4HVZ4 B4PDT2 A0A067RJF6 A0A1A9ZFV2
PDB
2D4W
E-value=1.84645e-123,
Score=1134
Ontologies
PATHWAY
GO
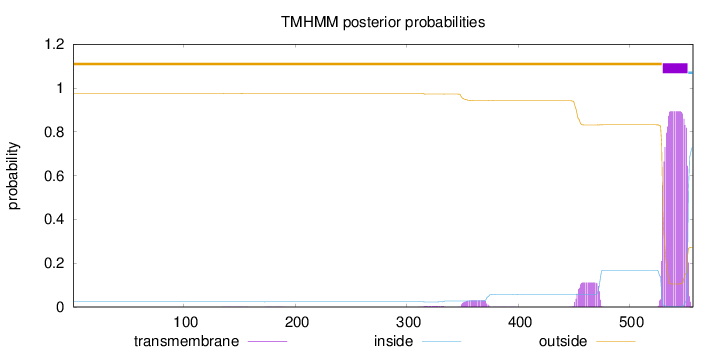
Topology
Length:
557
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.79703
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02431
outside
1 - 529
TMhelix
530 - 552
inside
553 - 557
Population Genetic Test Statistics
Pi
268.386764
Theta
180.769284
Tajima's D
1.550504
CLR
0.076584
CSRT
0.7977601119944
Interpretation
Uncertain
