Pre Gene Modal
BGIBMGA008203
Annotation
PREDICTED:_prostamide/prostaglandin_F_synthase-like_[Amyelois_transitella]
Full name
Prostamide/prostaglandin F synthase
Alternative Name
Peroxiredoxin-like 2B
Protein FAM213B
Protein FAM213B
Location in the cell
Cytoplasmic Reliability : 1.931
Sequence
CDS
ATGTCTCCGGATATTAATCAGATCGGTACGCAAAAAGTGAGAAACGTTTCCAGTGGCGAAGTTTTAGAATTGAAGAGCTTCTGGGAGCAACAGAATGTGGCTATTGTATTCTTTCGTCGCTGGGGTTGCATGTTTTGTAGGCTCTGGGCTAAAGAGTTGAGCGAAATAGCGCCTATTCTCAAGCAGCACAACATCAAACTTGTCGGGGTTGGCGTCGAAGAGGCCGGATCCAAGGAGTTCAGCGAAGGAAAGTTTTTCGATGGAGATCTGTATTACGTCGAGAACATTTCAACTTACCAACAGTTGGGTTTTAAAAGGTTCAATGTTTTAACTATTTTAACTTCGCTGCTATGGAAGCAATCACGTGACGCCATTTCGAAAGGAAAAGCCATGGGCTTAAGTGGCGACATGAAGGGAGATTGGGTTCAAGTGGGTGGCGCCGCTCTGGTAGAAAAGGGCGGGAACTTGCTCCGCCATTTTGTGCAAACCGGACCGGCGGATCATCTGTCTAATTTGGAGATTTTGAAGCAATTCGGTTTGGAAAATGAGTACAAAAAAGATGCGGAGAACGAAAAGAATTCGAAGAAAAACGAAGACACCACAGAGACAGAAAAAGAGTAA
Protein
MSPDINQIGTQKVRNVSSGEVLELKSFWEQQNVAIVFFRRWGCMFCRLWAKELSEIAPILKQHNIKLVGVGVEEAGSKEFSEGKFFDGDLYYVENISTYQQLGFKRFNVLTILTSLLWKQSRDAISKGKAMGLSGDMKGDWVQVGGAALVEKGGNLLRHFVQTGPADHLSNLEILKQFGLENEYKKDAENEKNSKKNEDTTETEKE
Summary
Description
Catalyzes the reduction of prostaglandin-ethanolamide H(2) (prostamide H(2)) to prostamide F(2alpha) with NADPH as proton donor. Also able to reduce prostaglandin H(2) to prostaglandin F(2alpha).
Catalyzes the reduction of prostaglandin-ethanolamide H(2) (prostamide H(2)) to prostamide F(2alpha) with NADPH as proton donor. Also able to reduce prostaglandin H(2) to prostaglandin F(2alpha) (By similarity).
Catalyzes the reduction of prostaglandin-ethanolamide H(2) (prostamide H(2)) to prostamide F(2alpha) with NADPH as proton donor. Also able to reduce prostaglandin H(2) to prostaglandin F(2alpha) (By similarity).
Catalytic Activity
[thioredoxin]-dithiol + prostaglandin H2 = [thioredoxin]-disulfide + prostaglandin F2alpha
[thioredoxin]-disulfide + prostamide F2alpha = [thioredoxin]-dithiol + prostamide H2
[thioredoxin]-disulfide + prostamide F2alpha = [thioredoxin]-dithiol + prostamide H2
Biophysicochemical Properties
0.0076 mM for prostamide H(2)
0.0069 mM for prostaglandin H(2)
0.0069 mM for prostaglandin H(2)
Similarity
Belongs to the peroxiredoxin-like PRXL2 family. Prostamide/prostaglandin F synthase subfamily.
Keywords
Complete proteome
Cytoplasm
Fatty acid biosynthesis
Fatty acid metabolism
Lipid biosynthesis
Lipid metabolism
NADP
Oxidoreductase
Phosphoprotein
Prostaglandin biosynthesis
Prostaglandin metabolism
Reference proteome
Alternative splicing
Feature
chain Prostamide/prostaglandin F synthase
splice variant In isoform 3 and isoform 6.
splice variant In isoform 3 and isoform 6.
Uniprot
H9JFA6
A0A2W1BDD1
A0A194Q702
A0A1E1WCC6
I4DR63
A0A3S2PFG0
+ More
A0A212EJ08 A0A0N0PDA8 A0A1B6D4E4 A0A2H1WBS8 A0A1B6M9A7 A0A1B6EYU7 A0A1B6JQX3 F6ZXR1 A0A250YBW7 C3ZVQ0 A0A1S3A5B3 A0A3B4ZMU3 Q9DB60 A0A1S3WJN1 H0WBZ1 G3TNZ7 A0A340WPL8 A0A2Y9SXY0 Q28IJ3 A0A2Y9DZS0 D3ZVR7 A0A2Y9Q9B4 A0A2J8URL9 A0A3P9AP83 A0A2Y9QFP9 H0XJV3 Q8TBF2 A0A2P0CTB1 A0A3Q1BTG3 A0A3Q1GKI4 A0A2K6CY94 A0A2K5MX57 A0A1B1V5U4 A9CQL8 A0A1U7R4K1 H2PXV2 L8YGP9 Q6NV24 F1QSW9 F7G9T2 A0A2I3RSW9 W5M990 Q5R7S9 A0A2K5WTM5 G3IFJ9 A0A3P8X663 A0A3P8S336 A0A140T841 Q58CY6 A0A3Q3GM42 A0A2Y9H5R5 M3WXY8 A0A1L8FMM1 E6ZFD0 Q6AZG8 A0A3Q7SY94 K7F8H6 G9L3X6 G1M3W1 M3Y824 A0A3P9NFK6 A0A3B4WL33 A0A3P9NFR9 A0A0D9S8V1 A0A1S3P2L7 B5X9L9 H3BI27 A0A3P4M0C4 A0A3B4WW83 A0A087Y8D4 A0A1A8GDV6 A0A1A8C0J3 A0A1A8V6K5 A0A3B4UQL3 W4YIU2 A0A060WEU3 A0A384DGN9 A0A2Y9KFW8 A0A2U3XLX2 A0A1A8QGQ2 A0A1A8KWE0 A0A1A8N088 A0A3B3YQU0 A0A3P9I499 E2QXW7 A0A0E9X3R6 A0A2U9BF24 A0A2U3WFJ1 A0A3B4C9G3 A0A3B4GQT0 A0A3P8Q1F7 A0A3P9C9S6
A0A212EJ08 A0A0N0PDA8 A0A1B6D4E4 A0A2H1WBS8 A0A1B6M9A7 A0A1B6EYU7 A0A1B6JQX3 F6ZXR1 A0A250YBW7 C3ZVQ0 A0A1S3A5B3 A0A3B4ZMU3 Q9DB60 A0A1S3WJN1 H0WBZ1 G3TNZ7 A0A340WPL8 A0A2Y9SXY0 Q28IJ3 A0A2Y9DZS0 D3ZVR7 A0A2Y9Q9B4 A0A2J8URL9 A0A3P9AP83 A0A2Y9QFP9 H0XJV3 Q8TBF2 A0A2P0CTB1 A0A3Q1BTG3 A0A3Q1GKI4 A0A2K6CY94 A0A2K5MX57 A0A1B1V5U4 A9CQL8 A0A1U7R4K1 H2PXV2 L8YGP9 Q6NV24 F1QSW9 F7G9T2 A0A2I3RSW9 W5M990 Q5R7S9 A0A2K5WTM5 G3IFJ9 A0A3P8X663 A0A3P8S336 A0A140T841 Q58CY6 A0A3Q3GM42 A0A2Y9H5R5 M3WXY8 A0A1L8FMM1 E6ZFD0 Q6AZG8 A0A3Q7SY94 K7F8H6 G9L3X6 G1M3W1 M3Y824 A0A3P9NFK6 A0A3B4WL33 A0A3P9NFR9 A0A0D9S8V1 A0A1S3P2L7 B5X9L9 H3BI27 A0A3P4M0C4 A0A3B4WW83 A0A087Y8D4 A0A1A8GDV6 A0A1A8C0J3 A0A1A8V6K5 A0A3B4UQL3 W4YIU2 A0A060WEU3 A0A384DGN9 A0A2Y9KFW8 A0A2U3XLX2 A0A1A8QGQ2 A0A1A8KWE0 A0A1A8N088 A0A3B3YQU0 A0A3P9I499 E2QXW7 A0A0E9X3R6 A0A2U9BF24 A0A2U3WFJ1 A0A3B4C9G3 A0A3B4GQT0 A0A3P8Q1F7 A0A3P9C9S6
EC Number
1.11.1.20
Pubmed
19121390
28756777
26354079
22651552
22118469
17495919
+ More
28087693 18563158 16141072 19468303 15489334 18006499 21183079 20950588 21993624 25069045 14702039 16710414 23186163 16136131 23385571 23594743 17431167 21804562 29704459 24487278 19393038 16305752 17975172 27762356 17381049 23236062 20010809 20433749 9215903 24755649 24813606 17554307 16341006 25613341 25186727
28087693 18563158 16141072 19468303 15489334 18006499 21183079 20950588 21993624 25069045 14702039 16710414 23186163 16136131 23385571 23594743 17431167 21804562 29704459 24487278 19393038 16305752 17975172 27762356 17381049 23236062 20010809 20433749 9215903 24755649 24813606 17554307 16341006 25613341 25186727
EMBL
BABH01027029
BABH01027030
KZ150334
PZC71337.1
KQ459386
KPJ01159.1
+ More
GDQN01006428 JAT84626.1 AK404961 BAM20403.1 RSAL01000055 RVE49907.1 AGBW02014547 OWR41450.1 KQ460254 KPJ16341.1 GEDC01016853 JAS20445.1 ODYU01007591 SOQ50497.1 GEBQ01029007 GEBQ01028018 GEBQ01022495 GEBQ01011608 GEBQ01007478 JAT10970.1 JAT11959.1 JAT17482.1 JAT28369.1 JAT32499.1 GECZ01026553 JAS43216.1 GECU01028505 GECU01006062 JAS79201.1 JAT01645.1 GFFW01003657 JAV41131.1 GG666691 EEN43392.1 AK005188 AL607032 BC030453 AAKN02055852 CR760360 CH473968 NDHI03003447 PNJ47890.1 AAQR03101132 AK057027 AK075273 AK291908 AK298926 AK303504 AK316243 AF425266 AL139246 CH471183 BC022547 KX444503 ANW12370.1 AB329665 AACZ04062807 GABD01005988 NBAG03000385 JAA27112.1 PNI31418.1 KB359209 ELV14245.1 BC068342 AL954137 JSUE03000068 AHAT01009756 AHAT01009757 CR860030 AQIA01006217 JH002406 RAZU01000081 EGW13170.1 RLQ74836.1 BT021811 BC114900 AANG04003082 CM004478 OCT72846.1 FQ310506 CBN80880.1 BC078028 AGCU01097725 JP023260 AES11858.1 ACTA01020114 AEYP01070627 AEYP01070628 AQIB01136152 BT047738 AFYH01000508 AFYH01000509 AFYH01000510 AFYH01000511 CYRY02004139 VCW68671.1 AYCK01000386 HAEB01021366 HAEC01001853 SBQ69930.1 HADZ01008440 HAEA01015856 SBP72381.1 HADY01016334 HAEJ01014979 SBS55436.1 AAGJ04047665 FR904518 CDQ65788.1 HAEH01011469 HAEI01014218 SBR92309.1 HAED01017636 HAEE01016672 SBR36722.1 HAEF01021344 HAEG01006332 SBR62503.1 AAEX03003869 GBXM01011451 JAH97126.1 CP026248 AWP02605.1
GDQN01006428 JAT84626.1 AK404961 BAM20403.1 RSAL01000055 RVE49907.1 AGBW02014547 OWR41450.1 KQ460254 KPJ16341.1 GEDC01016853 JAS20445.1 ODYU01007591 SOQ50497.1 GEBQ01029007 GEBQ01028018 GEBQ01022495 GEBQ01011608 GEBQ01007478 JAT10970.1 JAT11959.1 JAT17482.1 JAT28369.1 JAT32499.1 GECZ01026553 JAS43216.1 GECU01028505 GECU01006062 JAS79201.1 JAT01645.1 GFFW01003657 JAV41131.1 GG666691 EEN43392.1 AK005188 AL607032 BC030453 AAKN02055852 CR760360 CH473968 NDHI03003447 PNJ47890.1 AAQR03101132 AK057027 AK075273 AK291908 AK298926 AK303504 AK316243 AF425266 AL139246 CH471183 BC022547 KX444503 ANW12370.1 AB329665 AACZ04062807 GABD01005988 NBAG03000385 JAA27112.1 PNI31418.1 KB359209 ELV14245.1 BC068342 AL954137 JSUE03000068 AHAT01009756 AHAT01009757 CR860030 AQIA01006217 JH002406 RAZU01000081 EGW13170.1 RLQ74836.1 BT021811 BC114900 AANG04003082 CM004478 OCT72846.1 FQ310506 CBN80880.1 BC078028 AGCU01097725 JP023260 AES11858.1 ACTA01020114 AEYP01070627 AEYP01070628 AQIB01136152 BT047738 AFYH01000508 AFYH01000509 AFYH01000510 AFYH01000511 CYRY02004139 VCW68671.1 AYCK01000386 HAEB01021366 HAEC01001853 SBQ69930.1 HADZ01008440 HAEA01015856 SBP72381.1 HADY01016334 HAEJ01014979 SBS55436.1 AAGJ04047665 FR904518 CDQ65788.1 HAEH01011469 HAEI01014218 SBR92309.1 HAED01017636 HAEE01016672 SBR36722.1 HAEF01021344 HAEG01006332 SBR62503.1 AAEX03003869 GBXM01011451 JAH97126.1 CP026248 AWP02605.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000007151
UP000053240
UP000002280
+ More
UP000001554 UP000079721 UP000261400 UP000000589 UP000005447 UP000007646 UP000265300 UP000248484 UP000008143 UP000248480 UP000002494 UP000248483 UP000265140 UP000005225 UP000005640 UP000257160 UP000257200 UP000233120 UP000233060 UP000008227 UP000189706 UP000002277 UP000011518 UP000000437 UP000006718 UP000018468 UP000001595 UP000233100 UP000001075 UP000273346 UP000265120 UP000265080 UP000009136 UP000261660 UP000248481 UP000011712 UP000186698 UP000286640 UP000007267 UP000008912 UP000000715 UP000242638 UP000261360 UP000029965 UP000087266 UP000008672 UP000028760 UP000261420 UP000007110 UP000193380 UP000261680 UP000291021 UP000248482 UP000245341 UP000261480 UP000265200 UP000002254 UP000246464 UP000245340 UP000261440 UP000261460 UP000265100 UP000265160
UP000001554 UP000079721 UP000261400 UP000000589 UP000005447 UP000007646 UP000265300 UP000248484 UP000008143 UP000248480 UP000002494 UP000248483 UP000265140 UP000005225 UP000005640 UP000257160 UP000257200 UP000233120 UP000233060 UP000008227 UP000189706 UP000002277 UP000011518 UP000000437 UP000006718 UP000018468 UP000001595 UP000233100 UP000001075 UP000273346 UP000265120 UP000265080 UP000009136 UP000261660 UP000248481 UP000011712 UP000186698 UP000286640 UP000007267 UP000008912 UP000000715 UP000242638 UP000261360 UP000029965 UP000087266 UP000008672 UP000028760 UP000261420 UP000007110 UP000193380 UP000261680 UP000291021 UP000248482 UP000245341 UP000261480 UP000265200 UP000002254 UP000246464 UP000245340 UP000261440 UP000261460 UP000265100 UP000265160
Interpro
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
H9JFA6
A0A2W1BDD1
A0A194Q702
A0A1E1WCC6
I4DR63
A0A3S2PFG0
+ More
A0A212EJ08 A0A0N0PDA8 A0A1B6D4E4 A0A2H1WBS8 A0A1B6M9A7 A0A1B6EYU7 A0A1B6JQX3 F6ZXR1 A0A250YBW7 C3ZVQ0 A0A1S3A5B3 A0A3B4ZMU3 Q9DB60 A0A1S3WJN1 H0WBZ1 G3TNZ7 A0A340WPL8 A0A2Y9SXY0 Q28IJ3 A0A2Y9DZS0 D3ZVR7 A0A2Y9Q9B4 A0A2J8URL9 A0A3P9AP83 A0A2Y9QFP9 H0XJV3 Q8TBF2 A0A2P0CTB1 A0A3Q1BTG3 A0A3Q1GKI4 A0A2K6CY94 A0A2K5MX57 A0A1B1V5U4 A9CQL8 A0A1U7R4K1 H2PXV2 L8YGP9 Q6NV24 F1QSW9 F7G9T2 A0A2I3RSW9 W5M990 Q5R7S9 A0A2K5WTM5 G3IFJ9 A0A3P8X663 A0A3P8S336 A0A140T841 Q58CY6 A0A3Q3GM42 A0A2Y9H5R5 M3WXY8 A0A1L8FMM1 E6ZFD0 Q6AZG8 A0A3Q7SY94 K7F8H6 G9L3X6 G1M3W1 M3Y824 A0A3P9NFK6 A0A3B4WL33 A0A3P9NFR9 A0A0D9S8V1 A0A1S3P2L7 B5X9L9 H3BI27 A0A3P4M0C4 A0A3B4WW83 A0A087Y8D4 A0A1A8GDV6 A0A1A8C0J3 A0A1A8V6K5 A0A3B4UQL3 W4YIU2 A0A060WEU3 A0A384DGN9 A0A2Y9KFW8 A0A2U3XLX2 A0A1A8QGQ2 A0A1A8KWE0 A0A1A8N088 A0A3B3YQU0 A0A3P9I499 E2QXW7 A0A0E9X3R6 A0A2U9BF24 A0A2U3WFJ1 A0A3B4C9G3 A0A3B4GQT0 A0A3P8Q1F7 A0A3P9C9S6
A0A212EJ08 A0A0N0PDA8 A0A1B6D4E4 A0A2H1WBS8 A0A1B6M9A7 A0A1B6EYU7 A0A1B6JQX3 F6ZXR1 A0A250YBW7 C3ZVQ0 A0A1S3A5B3 A0A3B4ZMU3 Q9DB60 A0A1S3WJN1 H0WBZ1 G3TNZ7 A0A340WPL8 A0A2Y9SXY0 Q28IJ3 A0A2Y9DZS0 D3ZVR7 A0A2Y9Q9B4 A0A2J8URL9 A0A3P9AP83 A0A2Y9QFP9 H0XJV3 Q8TBF2 A0A2P0CTB1 A0A3Q1BTG3 A0A3Q1GKI4 A0A2K6CY94 A0A2K5MX57 A0A1B1V5U4 A9CQL8 A0A1U7R4K1 H2PXV2 L8YGP9 Q6NV24 F1QSW9 F7G9T2 A0A2I3RSW9 W5M990 Q5R7S9 A0A2K5WTM5 G3IFJ9 A0A3P8X663 A0A3P8S336 A0A140T841 Q58CY6 A0A3Q3GM42 A0A2Y9H5R5 M3WXY8 A0A1L8FMM1 E6ZFD0 Q6AZG8 A0A3Q7SY94 K7F8H6 G9L3X6 G1M3W1 M3Y824 A0A3P9NFK6 A0A3B4WL33 A0A3P9NFR9 A0A0D9S8V1 A0A1S3P2L7 B5X9L9 H3BI27 A0A3P4M0C4 A0A3B4WW83 A0A087Y8D4 A0A1A8GDV6 A0A1A8C0J3 A0A1A8V6K5 A0A3B4UQL3 W4YIU2 A0A060WEU3 A0A384DGN9 A0A2Y9KFW8 A0A2U3XLX2 A0A1A8QGQ2 A0A1A8KWE0 A0A1A8N088 A0A3B3YQU0 A0A3P9I499 E2QXW7 A0A0E9X3R6 A0A2U9BF24 A0A2U3WFJ1 A0A3B4C9G3 A0A3B4GQT0 A0A3P8Q1F7 A0A3P9C9S6
PDB
4DZA
E-value=0.0875019,
Score=80
Ontologies
PATHWAY
GO
PANTHER
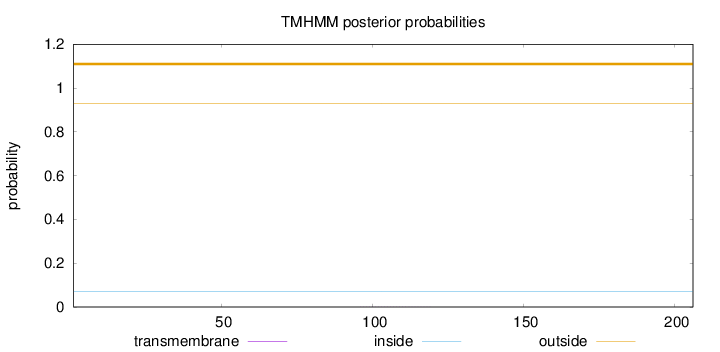
Topology
Subcellular location
Length:
206
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00919999999999998
Exp number, first 60 AAs:
0.00135
Total prob of N-in:
0.07060
outside
1 - 206
Population Genetic Test Statistics
Pi
224.439652
Theta
153.077834
Tajima's D
1.344527
CLR
0.741621
CSRT
0.754162291885406
Interpretation
Uncertain
