Gene
KWMTBOMO14340
Pre Gene Modal
BGIBMGA008194
Annotation
PREDICTED:_monoacylglycerol_lipase_ABHD12-like_isoform_X1_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 1.169
Sequence
CDS
ATGTTTGATTTGACCGGTATTTGTATTTTAGTGCCCCTTTATACAGCTGGAATACTGCTGCTTGGTGCAACAATAACGTCCGGAGTGTTCGTTTTCCACGTAGCTGTTGTACCACTAATATTCAAATATTCAAAATCATTTAGGAGGAATCTAGTCTTTGCTAATTTTGTACAATGGCCTCTGAATGTGAACTTCGAAGAACCCTCGTCAAGTGGTATAGAGGGCGGCAGGAATCTGAGCGTGGAGTATCATTCAAAAGTTGACAATTGTCCAATCAAAATAGGTATATGGCATATCCTACCGAGAACAAGCTATGAAAGATTGAAAGGAAATTTCGATGTTGATTCAGACAAGGAGGAATTAAATAGAATAATGGATCATGAATTGGCTACGTCCAAAACTCCAATAATGCTTTATTGCCACGGAAATTCAAATTCGCGCGCCGCCTCTCACAGAATACAGCTGTATAAGTTCTTTCAGAAAATGGATTTCCACACCATCGCTTTTGATTACAGAGGATATGGCGATTCCACGAACCTGTGTCCGACTGAGGAGGGCGTCGTGGAAGACGCACTGGTCGTCTACGACTGGCTTCTGGCCACCGCAGACAAGGGTGGGGAGACGCCGCCTGTATTCGTATGGGGACACTCTTTGGGAACGGCGATATCAGCGCACCTCCTCGGCAACTTGGAGCAGCTCTCGCTTCGGGTCCTGGGCCGCGCCCTGCCGCCGCCCCGCGGGCTCGTGCTGGAGGCGCCCTTCAACAACTTAGCGGATGAAGTAGCGAAGCACCCACTGTCCAAGCTGGTGACGTGGCTGCCGTACTACGAGGCTACCTTCGTGGAGCCCTTCAGGGTCCACCCCGAGCAGAGGTTCCAGTCTGAGAGGCACCTGGGTGGGGCCCCAGGGCTACCCGTGCTCATACTGCACGCCAGGGACGACGTCGTGGTGCCCTTCGCCGTCGGACTCCAGTTGTACAAATCGATACTAGCTTCGCGCTCGCAAGGTGGCGCCACCGTCGCGTTACATGCCTACGAGAAATCCGAGAACCTGGGCCACAAGTGGATATGCGAGGCCAAAGACTTGACCCAAGTTATAGGAGATTTCGTTACGGCTCATCTATGA
Protein
MFDLTGICILVPLYTAGILLLGATITSGVFVFHVAVVPLIFKYSKSFRRNLVFANFVQWPLNVNFEEPSSSGIEGGRNLSVEYHSKVDNCPIKIGIWHILPRTSYERLKGNFDVDSDKEELNRIMDHELATSKTPIMLYCHGNSNSRAASHRIQLYKFFQKMDFHTIAFDYRGYGDSTNLCPTEEGVVEDALVVYDWLLATADKGGETPPVFVWGHSLGTAISAHLLGNLEQLSLRVLGRALPPPRGLVLEAPFNNLADEVAKHPLSKLVTWLPYYEATFVEPFRVHPEQRFQSERHLGGAPGLPVLILHARDDVVVPFAVGLQLYKSILASRSQGGATVALHAYEKSENLGHKWICEAKDLTQVIGDFVTAHL
Summary
Uniprot
A0A2H1W3I5
A0A194Q858
A0A2A4JLD6
A0A2W1BHZ9
A0A212ELB9
S4P946
+ More
I4DPV0 Q5MGG1 A0A2W1BFR0 A0A0N1PI09 A0A2H1VJX8 A0A0L7RC58 A0A088AJ98 A0A2A3EUN9 E2C8H0 B0WF88 A0A232ELN2 K7IWE2 A0A023ERC5 A0A1Q3FNX9 A0A1Q3FP63 A0A1Q3FP04 A0A1Q3FNM2 Q170K7 A0A023ESN6 A0A1Q3FPE6 U5ETP8 A0A0V0GC71 A0A182QFV3 A0A084WUM0 A0NGJ1 A0A0J7KRM8 A0A310SF77 A0A1L8DXZ2 A0A224XN36 A0A1L8DY46 A0A1L8DYF3 A0A0K8TLP2 A0A182KSZ6 A0A182W296 A0A182PGE0 A0A182V0X1 A0A151IV80 A0A182RW65 A0A182TJ88 A0A1B0CAE4 A0A158NIJ4 A0A023F7G0 F4WPZ9 A0A182WZY5 A0A182JVM5 A0A182HPU3 A0A151K026 A0A195CJL1 A0A195BDX3 A0A2M3ZHD0 W5JTY7 A0A182YMJ2 A0A2M4AHX3 A0A2M4AFY6 A0A2M4AFT3 A0A1W4WJT1 A0A336L1B9 A0A336L132 A0A151XFW8 A0A336L4W6 E9I8E5 A0A1W4WUM3 N6TNN5 A0A0L0C1M8 A0A1W4WJU8 A0A1B6D3H0 A0A1B6D7Z0 E2A7B9 N6TRB6 A0A182MXA2 A0A0J9RGE2 A0A0P4WRE6 A0A1A9YDR9 A0A0Q5VM83 R4G5U5 T1H967 B4HPC1 A0A0R1DS30 B4QDM0 B3NKF4 A0A1A9ZCY2 A0A1B0GQ17 A0A067QHM6 A1ZBH2 B4PBP9 Q8IGV0 W8BWM7 W8C4W6 A0A2S2R2E5 W8CB11 A0A1B0AUW0 A1ZBH3
I4DPV0 Q5MGG1 A0A2W1BFR0 A0A0N1PI09 A0A2H1VJX8 A0A0L7RC58 A0A088AJ98 A0A2A3EUN9 E2C8H0 B0WF88 A0A232ELN2 K7IWE2 A0A023ERC5 A0A1Q3FNX9 A0A1Q3FP63 A0A1Q3FP04 A0A1Q3FNM2 Q170K7 A0A023ESN6 A0A1Q3FPE6 U5ETP8 A0A0V0GC71 A0A182QFV3 A0A084WUM0 A0NGJ1 A0A0J7KRM8 A0A310SF77 A0A1L8DXZ2 A0A224XN36 A0A1L8DY46 A0A1L8DYF3 A0A0K8TLP2 A0A182KSZ6 A0A182W296 A0A182PGE0 A0A182V0X1 A0A151IV80 A0A182RW65 A0A182TJ88 A0A1B0CAE4 A0A158NIJ4 A0A023F7G0 F4WPZ9 A0A182WZY5 A0A182JVM5 A0A182HPU3 A0A151K026 A0A195CJL1 A0A195BDX3 A0A2M3ZHD0 W5JTY7 A0A182YMJ2 A0A2M4AHX3 A0A2M4AFY6 A0A2M4AFT3 A0A1W4WJT1 A0A336L1B9 A0A336L132 A0A151XFW8 A0A336L4W6 E9I8E5 A0A1W4WUM3 N6TNN5 A0A0L0C1M8 A0A1W4WJU8 A0A1B6D3H0 A0A1B6D7Z0 E2A7B9 N6TRB6 A0A182MXA2 A0A0J9RGE2 A0A0P4WRE6 A0A1A9YDR9 A0A0Q5VM83 R4G5U5 T1H967 B4HPC1 A0A0R1DS30 B4QDM0 B3NKF4 A0A1A9ZCY2 A0A1B0GQ17 A0A067QHM6 A1ZBH2 B4PBP9 Q8IGV0 W8BWM7 W8C4W6 A0A2S2R2E5 W8CB11 A0A1B0AUW0 A1ZBH3
Pubmed
26354079
28756777
22118469
23622113
22651552
16023793
+ More
20798317 28648823 20075255 24945155 17510324 24438588 12364791 26369729 20966253 21347285 25474469 21719571 20920257 23761445 25244985 21282665 23537049 26108605 22936249 17994087 17550304 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 26109357 26109356
20798317 28648823 20075255 24945155 17510324 24438588 12364791 26369729 20966253 21347285 25474469 21719571 20920257 23761445 25244985 21282665 23537049 26108605 22936249 17994087 17550304 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 26109357 26109356
EMBL
ODYU01006104
SOQ47650.1
KQ459386
KPJ01175.1
NWSH01001059
PCG72867.1
+ More
KZ150079 PZC73901.1 AGBW02014096 OWR42268.1 GAIX01003889 JAA88671.1 AK403795 BAM19940.1 AY829825 AAV91439.1 PZC73902.1 KQ460601 KPJ13803.1 ODYU01002708 SOQ40554.1 KQ414617 KOC68408.1 KZ288185 PBC34992.1 GL453666 EFN75700.1 DS231915 EDS26064.1 NNAY01003522 OXU19269.1 GAPW01001868 JAC11730.1 GFDL01005862 JAV29183.1 GFDL01005769 JAV29276.1 GFDL01005781 JAV29264.1 GFDL01005943 JAV29102.1 CH477472 EAT40389.1 GAPW01001298 JAC12300.1 GFDL01005663 JAV29382.1 GANO01001809 JAB58062.1 GECL01000552 JAP05572.1 AXCN02001677 ATLV01027100 KE525423 KFB53914.1 AAAB01008986 EAU75801.2 LBMM01003959 KMQ92951.1 KQ769255 OAD52879.1 GFDF01002754 JAV11330.1 GFTR01005198 JAW11228.1 GFDF01002755 JAV11329.1 GFDF01002594 JAV11490.1 GDAI01002537 JAI15066.1 KQ980913 KYN11464.1 AJWK01003704 AJWK01003705 AJWK01003706 AJWK01003707 AJWK01003708 ADTU01016859 GBBI01001540 JAC17172.1 GL888261 EGI63728.1 APCN01003272 KQ981298 KYN43064.1 KQ977642 KYN00903.1 KQ976513 KYM82402.1 GGFM01007168 MBW27919.1 ADMH02000489 ETN66224.1 GGFK01007068 MBW40389.1 GGFK01006365 MBW39686.1 GGFK01006330 MBW39651.1 UFQS01001603 UFQT01001603 SSX11570.1 SSX31137.1 SSX11568.1 SSX31135.1 KQ982179 KYQ59273.1 SSX11569.1 SSX31136.1 GL761538 EFZ23140.1 APGK01057312 KB741280 KB631604 ENN70865.1 ERL84583.1 JRES01001007 KNC26176.1 GEDC01031521 GEDC01017049 JAS05777.1 JAS20249.1 GEDC01015532 GEDC01010329 GEDC01003706 JAS21766.1 JAS26969.1 JAS33592.1 GL437329 EFN70616.1 ENN70866.1 ERL84584.1 CM002911 KMY94991.1 GDRN01043356 JAI66986.1 CH954179 KQS62124.1 GAHY01000215 JAA77295.1 ACPB03001589 CH480816 EDW48559.1 CM000158 KRJ99938.1 KRJ99940.1 CM000362 EDX07776.1 KMY94990.1 EDV55176.1 AJVK01035450 AJVK01035451 AJVK01035452 KK853366 KDR08111.1 AE013599 AAF57599.2 EDW91533.1 KRJ99939.1 BT001585 AAN71340.1 GAMC01005247 JAC01309.1 GAMC01005249 GAMC01005248 JAC01307.1 GGMS01015008 MBY84211.1 GAMC01005246 JAC01310.1 JXJN01003752 BT050565 AAM68437.1 ACJ23449.1
KZ150079 PZC73901.1 AGBW02014096 OWR42268.1 GAIX01003889 JAA88671.1 AK403795 BAM19940.1 AY829825 AAV91439.1 PZC73902.1 KQ460601 KPJ13803.1 ODYU01002708 SOQ40554.1 KQ414617 KOC68408.1 KZ288185 PBC34992.1 GL453666 EFN75700.1 DS231915 EDS26064.1 NNAY01003522 OXU19269.1 GAPW01001868 JAC11730.1 GFDL01005862 JAV29183.1 GFDL01005769 JAV29276.1 GFDL01005781 JAV29264.1 GFDL01005943 JAV29102.1 CH477472 EAT40389.1 GAPW01001298 JAC12300.1 GFDL01005663 JAV29382.1 GANO01001809 JAB58062.1 GECL01000552 JAP05572.1 AXCN02001677 ATLV01027100 KE525423 KFB53914.1 AAAB01008986 EAU75801.2 LBMM01003959 KMQ92951.1 KQ769255 OAD52879.1 GFDF01002754 JAV11330.1 GFTR01005198 JAW11228.1 GFDF01002755 JAV11329.1 GFDF01002594 JAV11490.1 GDAI01002537 JAI15066.1 KQ980913 KYN11464.1 AJWK01003704 AJWK01003705 AJWK01003706 AJWK01003707 AJWK01003708 ADTU01016859 GBBI01001540 JAC17172.1 GL888261 EGI63728.1 APCN01003272 KQ981298 KYN43064.1 KQ977642 KYN00903.1 KQ976513 KYM82402.1 GGFM01007168 MBW27919.1 ADMH02000489 ETN66224.1 GGFK01007068 MBW40389.1 GGFK01006365 MBW39686.1 GGFK01006330 MBW39651.1 UFQS01001603 UFQT01001603 SSX11570.1 SSX31137.1 SSX11568.1 SSX31135.1 KQ982179 KYQ59273.1 SSX11569.1 SSX31136.1 GL761538 EFZ23140.1 APGK01057312 KB741280 KB631604 ENN70865.1 ERL84583.1 JRES01001007 KNC26176.1 GEDC01031521 GEDC01017049 JAS05777.1 JAS20249.1 GEDC01015532 GEDC01010329 GEDC01003706 JAS21766.1 JAS26969.1 JAS33592.1 GL437329 EFN70616.1 ENN70866.1 ERL84584.1 CM002911 KMY94991.1 GDRN01043356 JAI66986.1 CH954179 KQS62124.1 GAHY01000215 JAA77295.1 ACPB03001589 CH480816 EDW48559.1 CM000158 KRJ99938.1 KRJ99940.1 CM000362 EDX07776.1 KMY94990.1 EDV55176.1 AJVK01035450 AJVK01035451 AJVK01035452 KK853366 KDR08111.1 AE013599 AAF57599.2 EDW91533.1 KRJ99939.1 BT001585 AAN71340.1 GAMC01005247 JAC01309.1 GAMC01005249 GAMC01005248 JAC01307.1 GGMS01015008 MBY84211.1 GAMC01005246 JAC01310.1 JXJN01003752 BT050565 AAM68437.1 ACJ23449.1
Proteomes
UP000053268
UP000218220
UP000007151
UP000053240
UP000053825
UP000005203
+ More
UP000242457 UP000008237 UP000002320 UP000215335 UP000002358 UP000008820 UP000075886 UP000030765 UP000007062 UP000036403 UP000075882 UP000075920 UP000075885 UP000075903 UP000078492 UP000075900 UP000075902 UP000092461 UP000005205 UP000007755 UP000076407 UP000075881 UP000075840 UP000078541 UP000078542 UP000078540 UP000000673 UP000076408 UP000192223 UP000075809 UP000019118 UP000030742 UP000037069 UP000000311 UP000075884 UP000092443 UP000008711 UP000015103 UP000001292 UP000002282 UP000000304 UP000092445 UP000092462 UP000027135 UP000000803 UP000092460
UP000242457 UP000008237 UP000002320 UP000215335 UP000002358 UP000008820 UP000075886 UP000030765 UP000007062 UP000036403 UP000075882 UP000075920 UP000075885 UP000075903 UP000078492 UP000075900 UP000075902 UP000092461 UP000005205 UP000007755 UP000076407 UP000075881 UP000075840 UP000078541 UP000078542 UP000078540 UP000000673 UP000076408 UP000192223 UP000075809 UP000019118 UP000030742 UP000037069 UP000000311 UP000075884 UP000092443 UP000008711 UP000015103 UP000001292 UP000002282 UP000000304 UP000092445 UP000092462 UP000027135 UP000000803 UP000092460
Pfam
Interpro
IPR026605
ABHD12
+ More
IPR029058 AB_hydrolase
IPR000073 AB_hydrolase_1
IPR022742 Hydrolase_4
IPR002156 RNaseH_domain
IPR011047 Quinoprotein_ADH-like_supfam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR019775 WD40_repeat_CS
IPR003197 QCR7
IPR036544 QCR7_sf
IPR004143 BPL_LPL_catalytic
IPR004562 LipoylTrfase_LipoateP_Ligase
IPR029058 AB_hydrolase
IPR000073 AB_hydrolase_1
IPR022742 Hydrolase_4
IPR002156 RNaseH_domain
IPR011047 Quinoprotein_ADH-like_supfam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR019775 WD40_repeat_CS
IPR003197 QCR7
IPR036544 QCR7_sf
IPR004143 BPL_LPL_catalytic
IPR004562 LipoylTrfase_LipoateP_Ligase
Gene 3D
ProteinModelPortal
A0A2H1W3I5
A0A194Q858
A0A2A4JLD6
A0A2W1BHZ9
A0A212ELB9
S4P946
+ More
I4DPV0 Q5MGG1 A0A2W1BFR0 A0A0N1PI09 A0A2H1VJX8 A0A0L7RC58 A0A088AJ98 A0A2A3EUN9 E2C8H0 B0WF88 A0A232ELN2 K7IWE2 A0A023ERC5 A0A1Q3FNX9 A0A1Q3FP63 A0A1Q3FP04 A0A1Q3FNM2 Q170K7 A0A023ESN6 A0A1Q3FPE6 U5ETP8 A0A0V0GC71 A0A182QFV3 A0A084WUM0 A0NGJ1 A0A0J7KRM8 A0A310SF77 A0A1L8DXZ2 A0A224XN36 A0A1L8DY46 A0A1L8DYF3 A0A0K8TLP2 A0A182KSZ6 A0A182W296 A0A182PGE0 A0A182V0X1 A0A151IV80 A0A182RW65 A0A182TJ88 A0A1B0CAE4 A0A158NIJ4 A0A023F7G0 F4WPZ9 A0A182WZY5 A0A182JVM5 A0A182HPU3 A0A151K026 A0A195CJL1 A0A195BDX3 A0A2M3ZHD0 W5JTY7 A0A182YMJ2 A0A2M4AHX3 A0A2M4AFY6 A0A2M4AFT3 A0A1W4WJT1 A0A336L1B9 A0A336L132 A0A151XFW8 A0A336L4W6 E9I8E5 A0A1W4WUM3 N6TNN5 A0A0L0C1M8 A0A1W4WJU8 A0A1B6D3H0 A0A1B6D7Z0 E2A7B9 N6TRB6 A0A182MXA2 A0A0J9RGE2 A0A0P4WRE6 A0A1A9YDR9 A0A0Q5VM83 R4G5U5 T1H967 B4HPC1 A0A0R1DS30 B4QDM0 B3NKF4 A0A1A9ZCY2 A0A1B0GQ17 A0A067QHM6 A1ZBH2 B4PBP9 Q8IGV0 W8BWM7 W8C4W6 A0A2S2R2E5 W8CB11 A0A1B0AUW0 A1ZBH3
I4DPV0 Q5MGG1 A0A2W1BFR0 A0A0N1PI09 A0A2H1VJX8 A0A0L7RC58 A0A088AJ98 A0A2A3EUN9 E2C8H0 B0WF88 A0A232ELN2 K7IWE2 A0A023ERC5 A0A1Q3FNX9 A0A1Q3FP63 A0A1Q3FP04 A0A1Q3FNM2 Q170K7 A0A023ESN6 A0A1Q3FPE6 U5ETP8 A0A0V0GC71 A0A182QFV3 A0A084WUM0 A0NGJ1 A0A0J7KRM8 A0A310SF77 A0A1L8DXZ2 A0A224XN36 A0A1L8DY46 A0A1L8DYF3 A0A0K8TLP2 A0A182KSZ6 A0A182W296 A0A182PGE0 A0A182V0X1 A0A151IV80 A0A182RW65 A0A182TJ88 A0A1B0CAE4 A0A158NIJ4 A0A023F7G0 F4WPZ9 A0A182WZY5 A0A182JVM5 A0A182HPU3 A0A151K026 A0A195CJL1 A0A195BDX3 A0A2M3ZHD0 W5JTY7 A0A182YMJ2 A0A2M4AHX3 A0A2M4AFY6 A0A2M4AFT3 A0A1W4WJT1 A0A336L1B9 A0A336L132 A0A151XFW8 A0A336L4W6 E9I8E5 A0A1W4WUM3 N6TNN5 A0A0L0C1M8 A0A1W4WJU8 A0A1B6D3H0 A0A1B6D7Z0 E2A7B9 N6TRB6 A0A182MXA2 A0A0J9RGE2 A0A0P4WRE6 A0A1A9YDR9 A0A0Q5VM83 R4G5U5 T1H967 B4HPC1 A0A0R1DS30 B4QDM0 B3NKF4 A0A1A9ZCY2 A0A1B0GQ17 A0A067QHM6 A1ZBH2 B4PBP9 Q8IGV0 W8BWM7 W8C4W6 A0A2S2R2E5 W8CB11 A0A1B0AUW0 A1ZBH3
PDB
3C5V
E-value=0.03863,
Score=86
Ontologies
PANTHER
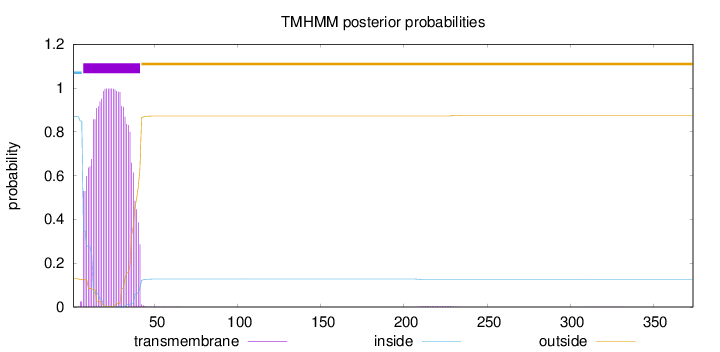
Topology
Length:
374
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
28.1576
Exp number, first 60 AAs:
28.08066
Total prob of N-in:
0.87023
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 41
outside
42 - 374
Population Genetic Test Statistics
Pi
293.05082
Theta
179.77684
Tajima's D
2.120431
CLR
0.098728
CSRT
0.900404979751012
Interpretation
Uncertain
