Gene
KWMTBOMO14338 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008126
Annotation
PREDICTED:_pro-resilin-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.895
Sequence
CDS
ATGGCGACGACGAATTCCAAAGATCTCTGTCTCAAGAGTACGGGGGTGTCCCAGGAATACGGTCTACCTAGCCGTAGTCTATCCCAAGAGTATGGAGCGCCTAAATCTCGATCAAATCTCAAACTTTCCCAAGACTACGGTCTACCTAACGCTCGTGCCAATTCAAAACCATCTCAAGTTTACGGTGCTCCTAATGCTCGTGGCTCTCCGTCTCTGGAATATGGACCTCCCCAAGACTTTTCCGGAGCGAACGTTGACTCTTATGCAGTGGAACATGCACCGTCATCTCAATATGGTACCCCTGAAGTACGTACACCTTCCGAGAATTACGGCGTGCCTCAAAGAAACCATGACCATCGCTCTCAGAAACAAGCCCGCAACCAAGCGTTATCTCGCAAATATGGTGTGCCTTCAGGCCGTTCGAACGACGTTTCTCAAAGTTACGGTGCTCCAAAGAACTCTTTCAAATCTTTCGCCGCCAGGAATTCACCTTCTCAAAGCTACGGCGCCCCTAAATCAGGACAGTTCGATGCTCCGTCCACTTCGTACGGTGTACCTAGTGCTTCTCAAGGATTCAAATCATTGTCTTCAACGTACGGTACTCCGAGTCAGAGAGGCTTGTCAGAGACATATGGAACTCCTTCCGCCAGGGGACTATCTCAAGAATACGGAGCTCCCACTGATTCTGAACTGAAGACTAAGGCTATTGCATCTACTTATGCATCTGGCAGAACGTCGCCTTCTCATGCCTATGGCGTACCGTCAGCGCGCGACTCCATGCCATCAGACCAGTACGGTATACCGGAACAATATTCGGCCCTAAACGACCAGAGCTATGAATACGCCAGAGCGGCTTTGGAAGAACTGAATCAGGAACCGGCTAATTACGATTTCGCTTACAAAGTAAACGACTATGTCAGCGGCAGTGACTTCGGACACGCGGAGTCGAGACAAGAGAATCGGGCCGAAGGAACGTATTTCGTGGTACTCCCAGATGGCACTAAACAGGTGGTAGAGTACGAAGCTGATGAGCGTGGCTTCAAGCCCAGGATCTCAGTGGAACCGGTCGAAACCGACCTTGGCTATGACGACAACGCATCAGATTTGACCAAATCTGAGAACGGTCCTTATTAG
Protein
MATTNSKDLCLKSTGVSQEYGLPSRSLSQEYGAPKSRSNLKLSQDYGLPNARANSKPSQVYGAPNARGSPSLEYGPPQDFSGANVDSYAVEHAPSSQYGTPEVRTPSENYGVPQRNHDHRSQKQARNQALSRKYGVPSGRSNDVSQSYGAPKNSFKSFAARNSPSQSYGAPKSGQFDAPSTSYGVPSASQGFKSLSSTYGTPSQRGLSETYGTPSARGLSQEYGAPTDSELKTKAIASTYASGRTSPSHAYGVPSARDSMPSDQYGIPEQYSALNDQSYEYARAALEELNQEPANYDFAYKVNDYVSGSDFGHAESRQENRAEGTYFVVLPDGTKQVVEYEADERGFKPRISVEPVETDLGYDDNASDLTKSENGPY
Summary
Uniprot
C0H6Y7
A0A3S2LBC5
A0A2A4JLA9
A0A2H1W490
A0A2W1BFL7
A0A194Q6L3
+ More
A0A212ELA1 A0A0N1IE56 A0A0N1PH11 A0A194Q6L6 A0A2R7WK82 A0A3S2L6J9 A0A2J7PLD3 A0A2R7WKQ1 A0A212EWL7 A0A2A4K9S1 A0A067R5G7 A0A2H1WG73 A0A2S1ZSD9 A0A2J7PLE2 A0A2P8YD37 A0A067QTK3 A0A2J7PLE5 A0A2J7PSE8 T1HNQ2 A0A0M4EKV5 A0A2R7WKE7 A0A2J7PLD7 A0A2R7WL13 A0A2P8YD20 B5DZE6 B4H740 T1HNQ3 A0A2S1ZSB2 A0A0J7KCH8 A0A2P8Z525 E0VR93 A0A182GQQ6 A0A2J7PLA7 A0A2J7PLA4 A0A067QS59 A0A182GZL4 A0A158NLC5 A0A195B2D2 A0A151IH02 A0A067R5G2 E2AT94 B4KSP6 A0A026WXZ7 A0A0L0CQL7 A0A195F1K7 A0A067QVX2 A0A182W2X6
A0A212ELA1 A0A0N1IE56 A0A0N1PH11 A0A194Q6L6 A0A2R7WK82 A0A3S2L6J9 A0A2J7PLD3 A0A2R7WKQ1 A0A212EWL7 A0A2A4K9S1 A0A067R5G7 A0A2H1WG73 A0A2S1ZSD9 A0A2J7PLE2 A0A2P8YD37 A0A067QTK3 A0A2J7PLE5 A0A2J7PSE8 T1HNQ2 A0A0M4EKV5 A0A2R7WKE7 A0A2J7PLD7 A0A2R7WL13 A0A2P8YD20 B5DZE6 B4H740 T1HNQ3 A0A2S1ZSB2 A0A0J7KCH8 A0A2P8Z525 E0VR93 A0A182GQQ6 A0A2J7PLA7 A0A2J7PLA4 A0A067QS59 A0A182GZL4 A0A158NLC5 A0A195B2D2 A0A151IH02 A0A067R5G2 E2AT94 B4KSP6 A0A026WXZ7 A0A0L0CQL7 A0A195F1K7 A0A067QVX2 A0A182W2X6
Pubmed
EMBL
BABH01026995
BR000646
FAA00648.1
RSAL01000055
RVE49919.1
NWSH01001059
+ More
PCG72865.1 ODYU01006104 SOQ47652.1 KZ150079 PZC73899.1 KQ459386 KPJ01177.1 AGBW02014096 OWR42266.1 KQ460601 KPJ13800.1 KPJ13799.1 KPJ01178.1 KK854974 PTY20054.1 RSAL01003663 RVE40248.1 NEVH01024426 PNF17144.1 PTY20049.1 AGBW02011958 OWR45869.1 NWSH01000023 PCG80648.1 KK852950 KDR13352.1 ODYU01008121 SOQ51454.1 MF942856 AWK28379.1 PNF17147.1 PYGN01000694 PSN42146.1 KDR13349.1 PNF17148.1 NEVH01021932 PNF19250.1 ACPB03007520 CP012524 ALC42092.1 PTY20048.1 PNF17146.1 PTY20051.1 PSN42147.1 CM000071 EDY69567.2 CH479216 EDW33573.1 MF942835 AWK28358.1 LBMM01009504 KMQ88078.1 PYGN01000192 PSN51590.1 DS235459 EEB15899.1 JXUM01081101 KQ563227 KXJ74251.1 PNF17108.1 PNF17110.1 KK853366 KDR08115.1 JXUM01099821 KQ564521 KXJ72142.1 ADTU01019320 KQ976662 KYM78440.1 KQ977647 KYN00845.1 KDR13347.1 GL442545 EFN63339.1 CH933808 EDW10545.2 KK107063 QOIP01000005 EZA60950.1 RLU22512.1 JRES01000062 KNC34472.1 KQ981864 KYN34353.1 KDR13351.1
PCG72865.1 ODYU01006104 SOQ47652.1 KZ150079 PZC73899.1 KQ459386 KPJ01177.1 AGBW02014096 OWR42266.1 KQ460601 KPJ13800.1 KPJ13799.1 KPJ01178.1 KK854974 PTY20054.1 RSAL01003663 RVE40248.1 NEVH01024426 PNF17144.1 PTY20049.1 AGBW02011958 OWR45869.1 NWSH01000023 PCG80648.1 KK852950 KDR13352.1 ODYU01008121 SOQ51454.1 MF942856 AWK28379.1 PNF17147.1 PYGN01000694 PSN42146.1 KDR13349.1 PNF17148.1 NEVH01021932 PNF19250.1 ACPB03007520 CP012524 ALC42092.1 PTY20048.1 PNF17146.1 PTY20051.1 PSN42147.1 CM000071 EDY69567.2 CH479216 EDW33573.1 MF942835 AWK28358.1 LBMM01009504 KMQ88078.1 PYGN01000192 PSN51590.1 DS235459 EEB15899.1 JXUM01081101 KQ563227 KXJ74251.1 PNF17108.1 PNF17110.1 KK853366 KDR08115.1 JXUM01099821 KQ564521 KXJ72142.1 ADTU01019320 KQ976662 KYM78440.1 KQ977647 KYN00845.1 KDR13347.1 GL442545 EFN63339.1 CH933808 EDW10545.2 KK107063 QOIP01000005 EZA60950.1 RLU22512.1 JRES01000062 KNC34472.1 KQ981864 KYN34353.1 KDR13351.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000053240
+ More
UP000235965 UP000027135 UP000245037 UP000015103 UP000092553 UP000001819 UP000008744 UP000036403 UP000009046 UP000069940 UP000249989 UP000005205 UP000078540 UP000078542 UP000000311 UP000009192 UP000053097 UP000279307 UP000037069 UP000078541 UP000075920
UP000235965 UP000027135 UP000245037 UP000015103 UP000092553 UP000001819 UP000008744 UP000036403 UP000009046 UP000069940 UP000249989 UP000005205 UP000078540 UP000078542 UP000000311 UP000009192 UP000053097 UP000279307 UP000037069 UP000078541 UP000075920
PRIDE
ProteinModelPortal
C0H6Y7
A0A3S2LBC5
A0A2A4JLA9
A0A2H1W490
A0A2W1BFL7
A0A194Q6L3
+ More
A0A212ELA1 A0A0N1IE56 A0A0N1PH11 A0A194Q6L6 A0A2R7WK82 A0A3S2L6J9 A0A2J7PLD3 A0A2R7WKQ1 A0A212EWL7 A0A2A4K9S1 A0A067R5G7 A0A2H1WG73 A0A2S1ZSD9 A0A2J7PLE2 A0A2P8YD37 A0A067QTK3 A0A2J7PLE5 A0A2J7PSE8 T1HNQ2 A0A0M4EKV5 A0A2R7WKE7 A0A2J7PLD7 A0A2R7WL13 A0A2P8YD20 B5DZE6 B4H740 T1HNQ3 A0A2S1ZSB2 A0A0J7KCH8 A0A2P8Z525 E0VR93 A0A182GQQ6 A0A2J7PLA7 A0A2J7PLA4 A0A067QS59 A0A182GZL4 A0A158NLC5 A0A195B2D2 A0A151IH02 A0A067R5G2 E2AT94 B4KSP6 A0A026WXZ7 A0A0L0CQL7 A0A195F1K7 A0A067QVX2 A0A182W2X6
A0A212ELA1 A0A0N1IE56 A0A0N1PH11 A0A194Q6L6 A0A2R7WK82 A0A3S2L6J9 A0A2J7PLD3 A0A2R7WKQ1 A0A212EWL7 A0A2A4K9S1 A0A067R5G7 A0A2H1WG73 A0A2S1ZSD9 A0A2J7PLE2 A0A2P8YD37 A0A067QTK3 A0A2J7PLE5 A0A2J7PSE8 T1HNQ2 A0A0M4EKV5 A0A2R7WKE7 A0A2J7PLD7 A0A2R7WL13 A0A2P8YD20 B5DZE6 B4H740 T1HNQ3 A0A2S1ZSB2 A0A0J7KCH8 A0A2P8Z525 E0VR93 A0A182GQQ6 A0A2J7PLA7 A0A2J7PLA4 A0A067QS59 A0A182GZL4 A0A158NLC5 A0A195B2D2 A0A151IH02 A0A067R5G2 E2AT94 B4KSP6 A0A026WXZ7 A0A0L0CQL7 A0A195F1K7 A0A067QVX2 A0A182W2X6
Ontologies
GO
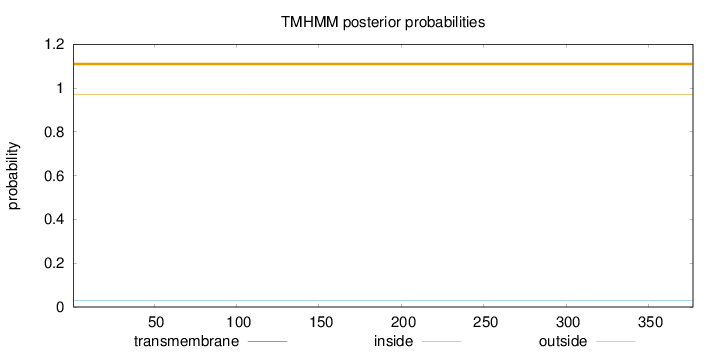
Topology
Length:
377
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03071
outside
1 - 377
Population Genetic Test Statistics
Pi
184.248689
Theta
28.919561
Tajima's D
0.54435
CLR
0.460401
CSRT
0.532573371331433
Interpretation
Uncertain
