Gene
KWMTBOMO14325 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008189
Annotation
enoyl-CoA_isomerase_[Biston_betularia]
Location in the cell
Cytoplasmic Reliability : 2.159
Sequence
CDS
ATGGCGGCGGAGGCTGAGGACCTTCCAGATGTGGCCAGGAAGGCCAGATACCATTACAACTTCATTAAATACGCACAGGACGGAATCACGGCTTTGGAGGATTGCGTCAAACCAGTGTTGACCGTGGTGCACAACGCCTGTATCGGCGCCGGAGTAGACCTGGTCACCGCCTGCGACATCAGGTATTGCTCCCAGGACGCGTGGTTTCAAGTCAAGGAAGTGGATGTTGGCTTGGCGGCCGATGTTGGAACCCTGCAAAGGCTCCCGAAGGTAATTGGCAACACGTCGATAGCGCGAGAACTTTGCTACACCGGCCGAAAGTTCGACGCCAGGGAAGCGAGTGAAATAGGCTTCGTCAGTAAAGTGTTCCCCGATAAAGACACTGCCCTCAGTCACGCGCTGCAGGTAGCCGAAGACGTCGCATCGAAAAGTCCGGTCGCAGTTCAAACGACGAAACAAAGTCTCGTCTACTCGCAGAGCAGACCAAACAGTGATGGCCTCGAGCATATTCGCTTGCTTAATTCCGTGATGAATCAAGGCGAGGATCTACCCAAAGCGGCCGTCGCTCAGGCCACGAAGAGCCCGCCGCCGGACTACGAGAACCTGTAG
Protein
MAAEAEDLPDVARKARYHYNFIKYAQDGITALEDCVKPVLTVVHNACIGAGVDLVTACDIRYCSQDAWFQVKEVDVGLAADVGTLQRLPKVIGNTSIARELCYTGRKFDAREASEIGFVSKVFPDKDTALSHALQVAEDVASKSPVAVQTTKQSLVYSQSRPNSDGLEHIRLLNSVMNQGEDLPKAAVAQATKSPPPDYENL
Summary
Similarity
Belongs to the enoyl-CoA hydratase/isomerase family.
Uniprot
H9JF92
G4XH86
A0A2H1VQ31
A0A2A4JB02
A0A068FLA1
A0A2W1BGJ6
+ More
A0A194Q6M3 A0A0L0C9M2 A0A1I8N316 A0A1B0AFE7 D3TS20 B4NE83 A0A1A9VNE5 B4M9X9 A0A2J7RLD2 A0A067QWT4 A0A1A9XKG4 A0A1B0AUD8 A0A0K8TN58 B3MQI8 Q29GV9 B4GV21 A0A1L8EH84 A0A1L8EGY6 A0A1L8EGU1 A0A1L8EH22 B4L7X6 U5EIN0 W8CB85 A0A3B0JSQ5 A0A1I8Q7B8 A0A336MTE1 A0A0M4FBI2 A0A1W4UQQ2 B4PYX4 A0A336MV42 Q9W5W8 A0A0K8WL63 A0A023F9K5 B4I645 A0A069DRC1 B3NWG3 A0A0V0G3Z2 A0A1W4XUG0 A0A0C9RFZ2 N6TYH4 V5GN47 A0A2S2PF08 A0A1L8DYW6 A0A034VU89 A0A0P4VV62 R4G4P5 A0A0K8UCS9 A0A0K8TWV3 A0A1W4XJ96 J3JWI2 A0A2S2PQ14 A0A023GI96 G3MNF2 A0A023GLB9 A0A2H8TV84 B4JK95 A0A0K8SQ84 A0A224YKX4 A0A1Q3FFF3 A0A1Q3FBP0 A0A1S4FW40 A0A1B6E6M7 Q16M31 A0A2P8Z477 A0A397W2Y2 A0A1S3K0E9 A0A023EMZ3 A0A1E1XA74 A0A182GHY2 E2C9X1 L7M4M3 A0A0C9RY96 A0A023FI95 A0A2S2QCY7 A0A0A9Y7G8 E0VFP2 A0A1B6FPH1 A0A1B6FJC8 L7LRY5 T1EMC1 A0A131YAY3 A0A195CQK2 A0A023EPE1 A0A0M9A0S2 C3YGP8 A0A0L7R934 A0A131YJS5 A0A293MD17 D6WH44 A0A131XCG8 A0A195F330 A0A158NZY9 A0A1B6KXK6 A0A1W3JNW4
A0A194Q6M3 A0A0L0C9M2 A0A1I8N316 A0A1B0AFE7 D3TS20 B4NE83 A0A1A9VNE5 B4M9X9 A0A2J7RLD2 A0A067QWT4 A0A1A9XKG4 A0A1B0AUD8 A0A0K8TN58 B3MQI8 Q29GV9 B4GV21 A0A1L8EH84 A0A1L8EGY6 A0A1L8EGU1 A0A1L8EH22 B4L7X6 U5EIN0 W8CB85 A0A3B0JSQ5 A0A1I8Q7B8 A0A336MTE1 A0A0M4FBI2 A0A1W4UQQ2 B4PYX4 A0A336MV42 Q9W5W8 A0A0K8WL63 A0A023F9K5 B4I645 A0A069DRC1 B3NWG3 A0A0V0G3Z2 A0A1W4XUG0 A0A0C9RFZ2 N6TYH4 V5GN47 A0A2S2PF08 A0A1L8DYW6 A0A034VU89 A0A0P4VV62 R4G4P5 A0A0K8UCS9 A0A0K8TWV3 A0A1W4XJ96 J3JWI2 A0A2S2PQ14 A0A023GI96 G3MNF2 A0A023GLB9 A0A2H8TV84 B4JK95 A0A0K8SQ84 A0A224YKX4 A0A1Q3FFF3 A0A1Q3FBP0 A0A1S4FW40 A0A1B6E6M7 Q16M31 A0A2P8Z477 A0A397W2Y2 A0A1S3K0E9 A0A023EMZ3 A0A1E1XA74 A0A182GHY2 E2C9X1 L7M4M3 A0A0C9RY96 A0A023FI95 A0A2S2QCY7 A0A0A9Y7G8 E0VFP2 A0A1B6FPH1 A0A1B6FJC8 L7LRY5 T1EMC1 A0A131YAY3 A0A195CQK2 A0A023EPE1 A0A0M9A0S2 C3YGP8 A0A0L7R934 A0A131YJS5 A0A293MD17 D6WH44 A0A131XCG8 A0A195F330 A0A158NZY9 A0A1B6KXK6 A0A1W3JNW4
Pubmed
19121390
26385554
28756777
26354079
26108605
25315136
+ More
20353571 17994087 24845553 26369729 15632085 24495485 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25474469 26334808 23537049 25348373 27129103 22516182 22216098 28797301 17510324 29403074 24945155 28503490 26483478 20798317 25576852 26131772 25401762 20566863 23254933 26830274 18563158 18362917 19820115 28049606 21347285
20353571 17994087 24845553 26369729 15632085 24495485 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25474469 26334808 23537049 25348373 27129103 22516182 22216098 28797301 17510324 29403074 24945155 28503490 26483478 20798317 25576852 26131772 25401762 20566863 23254933 26830274 18563158 18362917 19820115 28049606 21347285
EMBL
BABH01026955
JF811441
AEP43790.1
ODYU01003749
SOQ42908.1
NWSH01002191
+ More
PCG68946.1 KJ622113 AID66703.1 KZ150154 PZC72804.1 KQ459386 KPJ01187.1 JRES01000725 KNC28947.1 CCAG010015264 EZ424222 ADD20498.1 CH964239 EDW82052.2 CH940655 EDW66038.1 NEVH01002690 PNF41641.1 KK852863 KDR14796.1 JXJN01003606 GDAI01002243 JAI15360.1 CH902621 EDV44614.1 CH379064 EAL32000.1 CH479192 EDW26558.1 GFDG01000846 JAV17953.1 GFDG01000848 JAV17951.1 GFDG01000847 JAV17952.1 GFDG01000845 JAV17954.1 CH933814 EDW05551.1 GANO01002565 JAB57306.1 GAMC01000679 JAC05877.1 OUUW01000003 SPP78500.1 UFQS01002445 UFQT01002445 SSX14057.1 SSX33476.1 CP012528 ALC49982.1 CM000162 EDX02052.1 UFQS01001903 UFQT01001903 SSX12674.1 SSX32117.1 AE014298 AY071173 AAF45374.2 AAL48795.1 GDHF01022110 GDHF01001041 GDHF01000545 JAI30204.1 JAI51273.1 JAI51769.1 GBBI01001028 JAC17684.1 CH480823 EDW56251.1 GBGD01002459 JAC86430.1 CH954180 EDV46783.1 GECL01003324 JAP02800.1 GBYB01012062 JAG81829.1 APGK01050095 KB741165 ENN73426.1 GALX01005484 JAB62982.1 GGMR01014877 MBY27496.1 GFDF01002477 JAV11607.1 GAKP01012928 JAC46024.1 GDKW01000665 JAI55930.1 ACPB03002667 GAHY01000596 JAA76914.1 GDHF01027933 GDHF01018613 JAI24381.1 JAI33701.1 GDHF01033372 JAI18942.1 BT127600 AEE62562.1 GGMR01018769 MBY31388.1 GBBM01001792 JAC33626.1 JO843403 AEO35020.1 GBBM01001803 JAC33615.1 GFXV01006380 MBW18185.1 CH916370 EDV99997.1 GBRD01010845 GBRD01010844 GBRD01010843 GBRD01001031 JAG54980.1 GFPF01007151 MAA18297.1 GFDL01008771 JAV26274.1 GFDL01010078 JAV24967.1 GEDC01012557 GEDC01003710 JAS24741.1 JAS33588.1 CH477882 EAT35386.1 PYGN01000204 PSN51277.1 QKWP01000047 RIB29094.1 GAPW01003358 JAC10240.1 GFAC01003030 JAT96158.1 JXUM01064723 KQ562312 KXJ76177.1 GL453904 EFN75269.1 GACK01005753 JAA59281.1 GBZX01000059 JAG92681.1 GBBK01003016 JAC21466.1 GGMS01006395 MBY75598.1 GBHO01035334 GBHO01035332 GBHO01035331 GBHO01035330 GBHO01035328 GBHO01035327 GBHO01018109 GBHO01006011 JAG08270.1 JAG08272.1 JAG08273.1 JAG08274.1 JAG08276.1 JAG08277.1 JAG25495.1 JAG37593.1 DS235123 EEB12198.1 GECZ01017662 JAS52107.1 GECZ01019505 JAS50264.1 GACK01010549 JAA54485.1 AMQM01005962 KB097182 ESN98299.1 GEDV01012108 JAP76449.1 KQ977394 KYN02986.1 GAPW01002732 JAC10866.1 KQ435794 KOX74026.1 GG666512 EEN60558.1 KQ414628 KOC67338.1 GEDV01009360 JAP79197.1 GFWV01018889 MAA43617.1 KQ971330 EFA00967.2 GEFH01004791 JAP63790.1 KQ981856 KYN34587.1 ADTU01005136 GEBQ01023846 JAT16131.1
PCG68946.1 KJ622113 AID66703.1 KZ150154 PZC72804.1 KQ459386 KPJ01187.1 JRES01000725 KNC28947.1 CCAG010015264 EZ424222 ADD20498.1 CH964239 EDW82052.2 CH940655 EDW66038.1 NEVH01002690 PNF41641.1 KK852863 KDR14796.1 JXJN01003606 GDAI01002243 JAI15360.1 CH902621 EDV44614.1 CH379064 EAL32000.1 CH479192 EDW26558.1 GFDG01000846 JAV17953.1 GFDG01000848 JAV17951.1 GFDG01000847 JAV17952.1 GFDG01000845 JAV17954.1 CH933814 EDW05551.1 GANO01002565 JAB57306.1 GAMC01000679 JAC05877.1 OUUW01000003 SPP78500.1 UFQS01002445 UFQT01002445 SSX14057.1 SSX33476.1 CP012528 ALC49982.1 CM000162 EDX02052.1 UFQS01001903 UFQT01001903 SSX12674.1 SSX32117.1 AE014298 AY071173 AAF45374.2 AAL48795.1 GDHF01022110 GDHF01001041 GDHF01000545 JAI30204.1 JAI51273.1 JAI51769.1 GBBI01001028 JAC17684.1 CH480823 EDW56251.1 GBGD01002459 JAC86430.1 CH954180 EDV46783.1 GECL01003324 JAP02800.1 GBYB01012062 JAG81829.1 APGK01050095 KB741165 ENN73426.1 GALX01005484 JAB62982.1 GGMR01014877 MBY27496.1 GFDF01002477 JAV11607.1 GAKP01012928 JAC46024.1 GDKW01000665 JAI55930.1 ACPB03002667 GAHY01000596 JAA76914.1 GDHF01027933 GDHF01018613 JAI24381.1 JAI33701.1 GDHF01033372 JAI18942.1 BT127600 AEE62562.1 GGMR01018769 MBY31388.1 GBBM01001792 JAC33626.1 JO843403 AEO35020.1 GBBM01001803 JAC33615.1 GFXV01006380 MBW18185.1 CH916370 EDV99997.1 GBRD01010845 GBRD01010844 GBRD01010843 GBRD01001031 JAG54980.1 GFPF01007151 MAA18297.1 GFDL01008771 JAV26274.1 GFDL01010078 JAV24967.1 GEDC01012557 GEDC01003710 JAS24741.1 JAS33588.1 CH477882 EAT35386.1 PYGN01000204 PSN51277.1 QKWP01000047 RIB29094.1 GAPW01003358 JAC10240.1 GFAC01003030 JAT96158.1 JXUM01064723 KQ562312 KXJ76177.1 GL453904 EFN75269.1 GACK01005753 JAA59281.1 GBZX01000059 JAG92681.1 GBBK01003016 JAC21466.1 GGMS01006395 MBY75598.1 GBHO01035334 GBHO01035332 GBHO01035331 GBHO01035330 GBHO01035328 GBHO01035327 GBHO01018109 GBHO01006011 JAG08270.1 JAG08272.1 JAG08273.1 JAG08274.1 JAG08276.1 JAG08277.1 JAG25495.1 JAG37593.1 DS235123 EEB12198.1 GECZ01017662 JAS52107.1 GECZ01019505 JAS50264.1 GACK01010549 JAA54485.1 AMQM01005962 KB097182 ESN98299.1 GEDV01012108 JAP76449.1 KQ977394 KYN02986.1 GAPW01002732 JAC10866.1 KQ435794 KOX74026.1 GG666512 EEN60558.1 KQ414628 KOC67338.1 GEDV01009360 JAP79197.1 GFWV01018889 MAA43617.1 KQ971330 EFA00967.2 GEFH01004791 JAP63790.1 KQ981856 KYN34587.1 ADTU01005136 GEBQ01023846 JAT16131.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000037069
UP000095301
UP000092445
+ More
UP000092444 UP000007798 UP000078200 UP000008792 UP000235965 UP000027135 UP000092443 UP000092460 UP000007801 UP000001819 UP000008744 UP000009192 UP000268350 UP000095300 UP000092553 UP000192221 UP000002282 UP000000803 UP000001292 UP000008711 UP000192223 UP000019118 UP000015103 UP000001070 UP000008820 UP000245037 UP000266673 UP000085678 UP000069940 UP000249989 UP000008237 UP000009046 UP000015101 UP000078542 UP000053105 UP000001554 UP000053825 UP000007266 UP000078541 UP000005205
UP000092444 UP000007798 UP000078200 UP000008792 UP000235965 UP000027135 UP000092443 UP000092460 UP000007801 UP000001819 UP000008744 UP000009192 UP000268350 UP000095300 UP000092553 UP000192221 UP000002282 UP000000803 UP000001292 UP000008711 UP000192223 UP000019118 UP000015103 UP000001070 UP000008820 UP000245037 UP000266673 UP000085678 UP000069940 UP000249989 UP000008237 UP000009046 UP000015101 UP000078542 UP000053105 UP000001554 UP000053825 UP000007266 UP000078541 UP000005205
Pfam
PF00378 ECH_1
Interpro
SUPFAM
SSF52096
SSF52096
Gene 3D
ProteinModelPortal
H9JF92
G4XH86
A0A2H1VQ31
A0A2A4JB02
A0A068FLA1
A0A2W1BGJ6
+ More
A0A194Q6M3 A0A0L0C9M2 A0A1I8N316 A0A1B0AFE7 D3TS20 B4NE83 A0A1A9VNE5 B4M9X9 A0A2J7RLD2 A0A067QWT4 A0A1A9XKG4 A0A1B0AUD8 A0A0K8TN58 B3MQI8 Q29GV9 B4GV21 A0A1L8EH84 A0A1L8EGY6 A0A1L8EGU1 A0A1L8EH22 B4L7X6 U5EIN0 W8CB85 A0A3B0JSQ5 A0A1I8Q7B8 A0A336MTE1 A0A0M4FBI2 A0A1W4UQQ2 B4PYX4 A0A336MV42 Q9W5W8 A0A0K8WL63 A0A023F9K5 B4I645 A0A069DRC1 B3NWG3 A0A0V0G3Z2 A0A1W4XUG0 A0A0C9RFZ2 N6TYH4 V5GN47 A0A2S2PF08 A0A1L8DYW6 A0A034VU89 A0A0P4VV62 R4G4P5 A0A0K8UCS9 A0A0K8TWV3 A0A1W4XJ96 J3JWI2 A0A2S2PQ14 A0A023GI96 G3MNF2 A0A023GLB9 A0A2H8TV84 B4JK95 A0A0K8SQ84 A0A224YKX4 A0A1Q3FFF3 A0A1Q3FBP0 A0A1S4FW40 A0A1B6E6M7 Q16M31 A0A2P8Z477 A0A397W2Y2 A0A1S3K0E9 A0A023EMZ3 A0A1E1XA74 A0A182GHY2 E2C9X1 L7M4M3 A0A0C9RY96 A0A023FI95 A0A2S2QCY7 A0A0A9Y7G8 E0VFP2 A0A1B6FPH1 A0A1B6FJC8 L7LRY5 T1EMC1 A0A131YAY3 A0A195CQK2 A0A023EPE1 A0A0M9A0S2 C3YGP8 A0A0L7R934 A0A131YJS5 A0A293MD17 D6WH44 A0A131XCG8 A0A195F330 A0A158NZY9 A0A1B6KXK6 A0A1W3JNW4
A0A194Q6M3 A0A0L0C9M2 A0A1I8N316 A0A1B0AFE7 D3TS20 B4NE83 A0A1A9VNE5 B4M9X9 A0A2J7RLD2 A0A067QWT4 A0A1A9XKG4 A0A1B0AUD8 A0A0K8TN58 B3MQI8 Q29GV9 B4GV21 A0A1L8EH84 A0A1L8EGY6 A0A1L8EGU1 A0A1L8EH22 B4L7X6 U5EIN0 W8CB85 A0A3B0JSQ5 A0A1I8Q7B8 A0A336MTE1 A0A0M4FBI2 A0A1W4UQQ2 B4PYX4 A0A336MV42 Q9W5W8 A0A0K8WL63 A0A023F9K5 B4I645 A0A069DRC1 B3NWG3 A0A0V0G3Z2 A0A1W4XUG0 A0A0C9RFZ2 N6TYH4 V5GN47 A0A2S2PF08 A0A1L8DYW6 A0A034VU89 A0A0P4VV62 R4G4P5 A0A0K8UCS9 A0A0K8TWV3 A0A1W4XJ96 J3JWI2 A0A2S2PQ14 A0A023GI96 G3MNF2 A0A023GLB9 A0A2H8TV84 B4JK95 A0A0K8SQ84 A0A224YKX4 A0A1Q3FFF3 A0A1Q3FBP0 A0A1S4FW40 A0A1B6E6M7 Q16M31 A0A2P8Z477 A0A397W2Y2 A0A1S3K0E9 A0A023EMZ3 A0A1E1XA74 A0A182GHY2 E2C9X1 L7M4M3 A0A0C9RY96 A0A023FI95 A0A2S2QCY7 A0A0A9Y7G8 E0VFP2 A0A1B6FPH1 A0A1B6FJC8 L7LRY5 T1EMC1 A0A131YAY3 A0A195CQK2 A0A023EPE1 A0A0M9A0S2 C3YGP8 A0A0L7R934 A0A131YJS5 A0A293MD17 D6WH44 A0A131XCG8 A0A195F330 A0A158NZY9 A0A1B6KXK6 A0A1W3JNW4
PDB
1DCI
E-value=4.1844e-50,
Score=496
Ontologies
GO
Topology
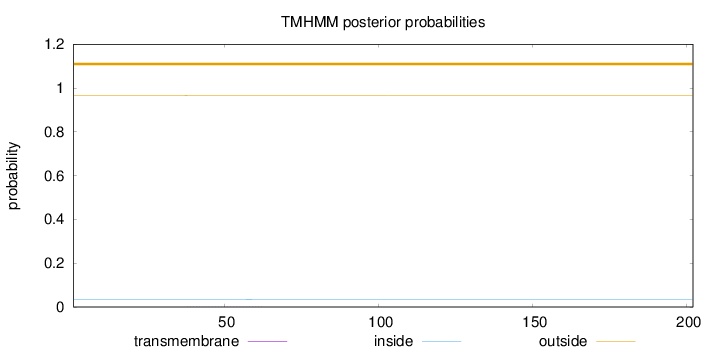
Length:
202
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01555
Exp number, first 60 AAs:
0.01504
Total prob of N-in:
0.03360
outside
1 - 202
Population Genetic Test Statistics
Pi
230.049472
Theta
152.320442
Tajima's D
1.642961
CLR
0.026582
CSRT
0.818309084545773
Interpretation
Uncertain
