Gene
KWMTBOMO14319
Pre Gene Modal
BGIBMGA008188
Annotation
PREDICTED:_probable_beta-hexosaminidase_fdl_[Bombyx_mori]
Full name
Beta-hexosaminidase
Location in the cell
PlasmaMembrane Reliability : 1.857
Sequence
CDS
ATGGGACCACAGCTTTGGCCTTACCCGATTGGCTATACGTTGTTCAGTAAAAATTTGGTAGCTATGTCGATCCTAAAATTGGAATACAAATTTCAATCGTTACCTTCAGAGAACGTGCATCGGCATTTAGCTGAAGCGTTTAAACTCTTCATTGGAGATTTAGCTCGATTGGAAAAGATAGGCAGAGAAATGAAAAACGGCACCTGTGATTTACCCGTCAAAAAGATGAATATTATTATAGATATCGAAACAGATCCCGATCCAAGATTGAGACTGAATACTGACGAAGCGTACACTATTAGTGTTAATAGCAATTTTGATTTTATTACCATACATTTAAGTTCCTCGTCTTTCTGTGGAGCTAGACATGGCTTGGAGACATTGAACCAGATGATTTTACTAGATCAACTGACCGGCTATTTGATAACTTTGTCGCATATTATTGTAAAGGACGCGCCAAGTCATAAATATAGAGGACTCATGATTGACACAGGAAGAAATTATATACCGATGCCAGATTTAATGAAAACTATTGATACAATGGCCGCTTGCAAATTAAATACATTTCACTGGAGGATATCCGATGTAACGAGCTTTCCGTTGCTTTTACCCAAGCTAAAAGAATTATTTGAGTTCGGTGCTTATGATAGAGCTTTGGTTTATACGAGAGCTGATGTGAAAGCTTTAGTGACGAGGGCTAAAGTTAGGGGGATTAGAGTATTGATTGAAGTGGCTGCACCGGGGCCGGTTGGACGACCTTGGTTTTGGCACAGCGAAGCGACTTGCCCGGCCAAAAATGCGAATTACACGTGCGATAACTTATTGTGTCTTAGATTAAATATGAGAGAGTCCATTTTTGATATATTACAATTAATATACGCCGAAATCATTGCACTCACTGAAGTGGACGATATATTCCATTTAAGTGATGGAGTATTCACAATGATCAATTGCTTTTCGTTGATTGAGGATAGAGATGGGTTTTTAGATAAAGCGCTGAATAGGCTAAGAATGGCCAATAAAGGCTTCATGCCGAAATTACCCATAGTCTGGTATACGAAACATCTGACCAGGACTCTTGAGGCGAAAACGTGGGAGCATTTGGGAGTGCAGCTTTATAAGTGGAACCCCGATTCTGGAGATTATTTGGGAACGTTTCGCGTGATTCATTCAAACAAATGGGATCTGTCGTGCGAAATGCGGAAACAGAGATGTTATAAATACCGGACTTGGCAAGGAATGTACTCGTGGACATCTTGGCGAAACATCGACGTGTTCACGATAGAAGGTGGAGAGGCAGTTCTTTGGACCGATTTTGTAGATTCAGGAAATCTTGATTATCACCTGTGGCCACGTGCTGCCGCAGTAGCCGAGAGACTGTGGTCAGATATAATAGCGAACACTACACCAAATCCTGAAGTCTACGTGAGATTAGATTACCATAGGTAA
Protein
MGPQLWPYPIGYTLFSKNLVAMSILKLEYKFQSLPSENVHRHLAEAFKLFIGDLARLEKIGREMKNGTCDLPVKKMNIIIDIETDPDPRLRLNTDEAYTISVNSNFDFITIHLSSSSFCGARHGLETLNQMILLDQLTGYLITLSHIIVKDAPSHKYRGLMIDTGRNYIPMPDLMKTIDTMAACKLNTFHWRISDVTSFPLLLPKLKELFEFGAYDRALVYTRADVKALVTRAKVRGIRVLIEVAAPGPVGRPWFWHSEATCPAKNANYTCDNLLCLRLNMRESIFDILQLIYAEIIALTEVDDIFHLSDGVFTMINCFSLIEDRDGFLDKALNRLRMANKGFMPKLPIVWYTKHLTRTLEAKTWEHLGVQLYKWNPDSGDYLGTFRVIHSNKWDLSCEMRKQRCYKYRTWQGMYSWTSWRNIDVFTIEGGEAVLWTDFVDSGNLDYHLWPRAAAVAERLWSDIIANTTPNPEVYVRLDYHR
Summary
Catalytic Activity
Hydrolysis of terminal non-reducing N-acetyl-D-hexosamine residues in N-acetyl-beta-D-hexosaminides.
Similarity
Belongs to the glycosyl hydrolase 20 family.
Feature
chain Beta-hexosaminidase
Uniprot
H9JF91
A0A2W1BL71
A0A0N1IH43
A0A0L7LDX4
A0A2A4ITA1
A0A212FFS2
+ More
A0A2H1WLD1 A0A194QCN4 B1P868 D6PW84 A0A194QCU8 A0A3S2NQV5 A0A212FNK4 A0A0L7LFG8 A0A0L7KQB6 D3GGI7 E1CG96 D3GGI8 D3GGI9 H9J3A9 A0A2A4JAM6 A0A212F886 A0A0L7LKV0 A0A194QZT5 A0A2A4JIQ5 A0A194QE79 A0A0G2RLW6 A0A194QYW8 A0A2H1VA55 A0A1Y1MUX2 A0A154PQQ9 A0A232F8H7 K7INU6 A0A0A7NXB7 A5YVX6 A0A0L7QN95 A0A2A3ES46 A0A088ANY0 A0A1L8E264 J3JWB4 U4UHA8 A0A026VXS5 F4WC87 A0A3L8D4R7 E2AN60 A0A2P8Y7F7 A0A1B0CI79 A0A195BK61 A0A195FQK5 A0A158N9Y2 A0A195C5C4 A0A151XJZ2 A0A1W4WN94 A0A1W4WN20 A0A1W4WM34 A0A1W4WC04 A0A1B0AU39 E2BWS6 A0A0T6BB39 A0A0J7LBD6 A0A1A9ZAJ7 A0A2J7REG4 A0A067QJM1 B4H544 A0A0R3NQ30 A0A0R3NWB6 A0A0R3NVQ4 Q28X77 A0A195EKI6 B3MEN3 A0A310SG90 A0A0Q9XGJ7 B4KPZ8 A0A3B0JFG0 A0A3B0JKP9 A0A1B1TKD7 A0A1A9V255 A0A1A9XPJ4 A0A1B0BXA1 U4UGG0 A0A1Q3F2V3 A0A1J1I7Y3 A0A1J1I989 A0A1Q3F2U8 A0A1B0FDX0 A5YVX5 D6WA88 A0A1A9X4S7 B4N602 A0A0L0C5J2 A0A182GY98 B0WNY9 A0A0M4E7X0 A0A0P6IVH7 A0A0R1DWG8 B4P590 A0A0R1DQK7 A0A0J9RB23 B4QCX6 A0A0J9RB31
A0A2H1WLD1 A0A194QCN4 B1P868 D6PW84 A0A194QCU8 A0A3S2NQV5 A0A212FNK4 A0A0L7LFG8 A0A0L7KQB6 D3GGI7 E1CG96 D3GGI8 D3GGI9 H9J3A9 A0A2A4JAM6 A0A212F886 A0A0L7LKV0 A0A194QZT5 A0A2A4JIQ5 A0A194QE79 A0A0G2RLW6 A0A194QYW8 A0A2H1VA55 A0A1Y1MUX2 A0A154PQQ9 A0A232F8H7 K7INU6 A0A0A7NXB7 A5YVX6 A0A0L7QN95 A0A2A3ES46 A0A088ANY0 A0A1L8E264 J3JWB4 U4UHA8 A0A026VXS5 F4WC87 A0A3L8D4R7 E2AN60 A0A2P8Y7F7 A0A1B0CI79 A0A195BK61 A0A195FQK5 A0A158N9Y2 A0A195C5C4 A0A151XJZ2 A0A1W4WN94 A0A1W4WN20 A0A1W4WM34 A0A1W4WC04 A0A1B0AU39 E2BWS6 A0A0T6BB39 A0A0J7LBD6 A0A1A9ZAJ7 A0A2J7REG4 A0A067QJM1 B4H544 A0A0R3NQ30 A0A0R3NWB6 A0A0R3NVQ4 Q28X77 A0A195EKI6 B3MEN3 A0A310SG90 A0A0Q9XGJ7 B4KPZ8 A0A3B0JFG0 A0A3B0JKP9 A0A1B1TKD7 A0A1A9V255 A0A1A9XPJ4 A0A1B0BXA1 U4UGG0 A0A1Q3F2V3 A0A1J1I7Y3 A0A1J1I989 A0A1Q3F2U8 A0A1B0FDX0 A5YVX5 D6WA88 A0A1A9X4S7 B4N602 A0A0L0C5J2 A0A182GY98 B0WNY9 A0A0M4E7X0 A0A0P6IVH7 A0A0R1DWG8 B4P590 A0A0R1DQK7 A0A0J9RB23 B4QCX6 A0A0J9RB31
EC Number
3.2.1.52
Pubmed
EMBL
BABH01026945
KZ149980
PZC75822.1
KQ460299
KPJ16149.1
JTDY01001544
+ More
KOB73584.1 NWSH01007588 PCG62899.1 AGBW02008773 OWR52577.1 ODYU01009424 SOQ53838.1 KQ459386 KPJ01201.1 EU334747 ODYU01005953 ACA30398.1 SOQ47378.1 GU984637 ADF97235.1 KQ459185 KPJ03244.1 RSAL01000352 RVE42263.1 AGBW02005277 OWR55318.1 JTDY01001324 KOB74197.1 JTDY01007043 KOB65498.1 FJ695479 ACV89845.1 AB540233 BAJ20189.1 FJ695480 ACV89846.1 BABH01021699 BABH01021700 FJ695481 ACV89847.1 BABH01021684 NWSH01002205 PCG68909.1 AGBW02009781 OWR49928.1 JTDY01000705 KOB76153.1 KQ460949 KPJ10490.1 NWSH01001413 PCG71313.1 KPJ03250.1 KP000851 AJG44546.1 KPJ10484.1 ODYU01001276 SOQ37272.1 GEZM01019981 JAV89483.1 KQ435007 KZC13460.1 NNAY01000660 OXU27126.1 KJ786476 AIZ97749.1 EF592539 KQ971354 ABQ95985.1 EFA06836.2 KQ414860 KOC60039.1 KZ288193 PBC34036.1 GFDF01001338 JAV12746.1 BT127532 AEE62494.1 KB632326 ERL92397.1 KK107796 EZA47639.1 GL888070 EGI68067.1 QOIP01000013 RLU15380.1 GL441088 EFN65124.1 PYGN01000845 PSN40104.1 AJWK01013083 KQ976455 KYM85038.1 KQ981305 KYN42860.1 ADTU01009908 KQ978292 KYM95386.1 KQ982052 KYQ60598.1 JXJN01003514 GL451188 EFN79790.1 LJIG01002462 KRT84478.1 LBMM01000051 KMR05153.1 NEVH01004981 PNF39232.1 KK853284 KDR09002.1 CH479210 EDW32880.1 CM000071 KRT03108.1 KRT03107.1 KRT03109.1 EAL26439.2 KQ978739 KYN28753.1 CH902619 EDV35497.2 KQ770827 OAD52570.1 CH933808 KRG03902.1 EDW08100.1 OUUW01000001 SPP74050.1 SPP74049.1 KU522433 ANV82809.1 JXJN01022146 KB632345 ERL93064.1 GFDL01013164 JAV21881.1 CVRI01000043 CRK96299.1 CRK96298.1 GFDL01013165 JAV21880.1 CCAG010006435 EF592538 ABQ95984.1 KQ971312 EEZ98597.1 CH964154 EDW79791.2 JRES01000960 KNC26699.1 JXUM01097258 JXUM01097259 JXUM01097260 JXUM01097261 DS232017 EDS32008.1 CP012524 ALC40696.1 GDUN01000092 JAN95827.1 CM000158 KRJ99443.1 EDW90752.2 KRJ99444.1 CM002911 KMY93226.1 CM000362 EDX06787.1 KMY93230.1 KMY93227.1
KOB73584.1 NWSH01007588 PCG62899.1 AGBW02008773 OWR52577.1 ODYU01009424 SOQ53838.1 KQ459386 KPJ01201.1 EU334747 ODYU01005953 ACA30398.1 SOQ47378.1 GU984637 ADF97235.1 KQ459185 KPJ03244.1 RSAL01000352 RVE42263.1 AGBW02005277 OWR55318.1 JTDY01001324 KOB74197.1 JTDY01007043 KOB65498.1 FJ695479 ACV89845.1 AB540233 BAJ20189.1 FJ695480 ACV89846.1 BABH01021699 BABH01021700 FJ695481 ACV89847.1 BABH01021684 NWSH01002205 PCG68909.1 AGBW02009781 OWR49928.1 JTDY01000705 KOB76153.1 KQ460949 KPJ10490.1 NWSH01001413 PCG71313.1 KPJ03250.1 KP000851 AJG44546.1 KPJ10484.1 ODYU01001276 SOQ37272.1 GEZM01019981 JAV89483.1 KQ435007 KZC13460.1 NNAY01000660 OXU27126.1 KJ786476 AIZ97749.1 EF592539 KQ971354 ABQ95985.1 EFA06836.2 KQ414860 KOC60039.1 KZ288193 PBC34036.1 GFDF01001338 JAV12746.1 BT127532 AEE62494.1 KB632326 ERL92397.1 KK107796 EZA47639.1 GL888070 EGI68067.1 QOIP01000013 RLU15380.1 GL441088 EFN65124.1 PYGN01000845 PSN40104.1 AJWK01013083 KQ976455 KYM85038.1 KQ981305 KYN42860.1 ADTU01009908 KQ978292 KYM95386.1 KQ982052 KYQ60598.1 JXJN01003514 GL451188 EFN79790.1 LJIG01002462 KRT84478.1 LBMM01000051 KMR05153.1 NEVH01004981 PNF39232.1 KK853284 KDR09002.1 CH479210 EDW32880.1 CM000071 KRT03108.1 KRT03107.1 KRT03109.1 EAL26439.2 KQ978739 KYN28753.1 CH902619 EDV35497.2 KQ770827 OAD52570.1 CH933808 KRG03902.1 EDW08100.1 OUUW01000001 SPP74050.1 SPP74049.1 KU522433 ANV82809.1 JXJN01022146 KB632345 ERL93064.1 GFDL01013164 JAV21881.1 CVRI01000043 CRK96299.1 CRK96298.1 GFDL01013165 JAV21880.1 CCAG010006435 EF592538 ABQ95984.1 KQ971312 EEZ98597.1 CH964154 EDW79791.2 JRES01000960 KNC26699.1 JXUM01097258 JXUM01097259 JXUM01097260 JXUM01097261 DS232017 EDS32008.1 CP012524 ALC40696.1 GDUN01000092 JAN95827.1 CM000158 KRJ99443.1 EDW90752.2 KRJ99444.1 CM002911 KMY93226.1 CM000362 EDX06787.1 KMY93230.1 KMY93227.1
Proteomes
UP000005204
UP000053240
UP000037510
UP000218220
UP000007151
UP000053268
+ More
UP000283053 UP000076502 UP000215335 UP000002358 UP000007266 UP000053825 UP000242457 UP000005203 UP000030742 UP000053097 UP000007755 UP000279307 UP000000311 UP000245037 UP000092461 UP000078540 UP000078541 UP000005205 UP000078542 UP000075809 UP000192223 UP000092460 UP000008237 UP000036403 UP000092445 UP000235965 UP000027135 UP000008744 UP000001819 UP000078492 UP000007801 UP000009192 UP000268350 UP000078200 UP000092443 UP000183832 UP000092444 UP000091820 UP000007798 UP000037069 UP000069940 UP000002320 UP000092553 UP000002282 UP000000304
UP000283053 UP000076502 UP000215335 UP000002358 UP000007266 UP000053825 UP000242457 UP000005203 UP000030742 UP000053097 UP000007755 UP000279307 UP000000311 UP000245037 UP000092461 UP000078540 UP000078541 UP000005205 UP000078542 UP000075809 UP000192223 UP000092460 UP000008237 UP000036403 UP000092445 UP000235965 UP000027135 UP000008744 UP000001819 UP000078492 UP000007801 UP000009192 UP000268350 UP000078200 UP000092443 UP000183832 UP000092444 UP000091820 UP000007798 UP000037069 UP000069940 UP000002320 UP000092553 UP000002282 UP000000304
Interpro
Gene 3D
ProteinModelPortal
H9JF91
A0A2W1BL71
A0A0N1IH43
A0A0L7LDX4
A0A2A4ITA1
A0A212FFS2
+ More
A0A2H1WLD1 A0A194QCN4 B1P868 D6PW84 A0A194QCU8 A0A3S2NQV5 A0A212FNK4 A0A0L7LFG8 A0A0L7KQB6 D3GGI7 E1CG96 D3GGI8 D3GGI9 H9J3A9 A0A2A4JAM6 A0A212F886 A0A0L7LKV0 A0A194QZT5 A0A2A4JIQ5 A0A194QE79 A0A0G2RLW6 A0A194QYW8 A0A2H1VA55 A0A1Y1MUX2 A0A154PQQ9 A0A232F8H7 K7INU6 A0A0A7NXB7 A5YVX6 A0A0L7QN95 A0A2A3ES46 A0A088ANY0 A0A1L8E264 J3JWB4 U4UHA8 A0A026VXS5 F4WC87 A0A3L8D4R7 E2AN60 A0A2P8Y7F7 A0A1B0CI79 A0A195BK61 A0A195FQK5 A0A158N9Y2 A0A195C5C4 A0A151XJZ2 A0A1W4WN94 A0A1W4WN20 A0A1W4WM34 A0A1W4WC04 A0A1B0AU39 E2BWS6 A0A0T6BB39 A0A0J7LBD6 A0A1A9ZAJ7 A0A2J7REG4 A0A067QJM1 B4H544 A0A0R3NQ30 A0A0R3NWB6 A0A0R3NVQ4 Q28X77 A0A195EKI6 B3MEN3 A0A310SG90 A0A0Q9XGJ7 B4KPZ8 A0A3B0JFG0 A0A3B0JKP9 A0A1B1TKD7 A0A1A9V255 A0A1A9XPJ4 A0A1B0BXA1 U4UGG0 A0A1Q3F2V3 A0A1J1I7Y3 A0A1J1I989 A0A1Q3F2U8 A0A1B0FDX0 A5YVX5 D6WA88 A0A1A9X4S7 B4N602 A0A0L0C5J2 A0A182GY98 B0WNY9 A0A0M4E7X0 A0A0P6IVH7 A0A0R1DWG8 B4P590 A0A0R1DQK7 A0A0J9RB23 B4QCX6 A0A0J9RB31
A0A2H1WLD1 A0A194QCN4 B1P868 D6PW84 A0A194QCU8 A0A3S2NQV5 A0A212FNK4 A0A0L7LFG8 A0A0L7KQB6 D3GGI7 E1CG96 D3GGI8 D3GGI9 H9J3A9 A0A2A4JAM6 A0A212F886 A0A0L7LKV0 A0A194QZT5 A0A2A4JIQ5 A0A194QE79 A0A0G2RLW6 A0A194QYW8 A0A2H1VA55 A0A1Y1MUX2 A0A154PQQ9 A0A232F8H7 K7INU6 A0A0A7NXB7 A5YVX6 A0A0L7QN95 A0A2A3ES46 A0A088ANY0 A0A1L8E264 J3JWB4 U4UHA8 A0A026VXS5 F4WC87 A0A3L8D4R7 E2AN60 A0A2P8Y7F7 A0A1B0CI79 A0A195BK61 A0A195FQK5 A0A158N9Y2 A0A195C5C4 A0A151XJZ2 A0A1W4WN94 A0A1W4WN20 A0A1W4WM34 A0A1W4WC04 A0A1B0AU39 E2BWS6 A0A0T6BB39 A0A0J7LBD6 A0A1A9ZAJ7 A0A2J7REG4 A0A067QJM1 B4H544 A0A0R3NQ30 A0A0R3NWB6 A0A0R3NVQ4 Q28X77 A0A195EKI6 B3MEN3 A0A310SG90 A0A0Q9XGJ7 B4KPZ8 A0A3B0JFG0 A0A3B0JKP9 A0A1B1TKD7 A0A1A9V255 A0A1A9XPJ4 A0A1B0BXA1 U4UGG0 A0A1Q3F2V3 A0A1J1I7Y3 A0A1J1I989 A0A1Q3F2U8 A0A1B0FDX0 A5YVX5 D6WA88 A0A1A9X4S7 B4N602 A0A0L0C5J2 A0A182GY98 B0WNY9 A0A0M4E7X0 A0A0P6IVH7 A0A0R1DWG8 B4P590 A0A0R1DQK7 A0A0J9RB23 B4QCX6 A0A0J9RB31
PDB
3S6T
E-value=1.65282e-45,
Score=461
Ontologies
KEGG
PATHWAY
GO
Topology
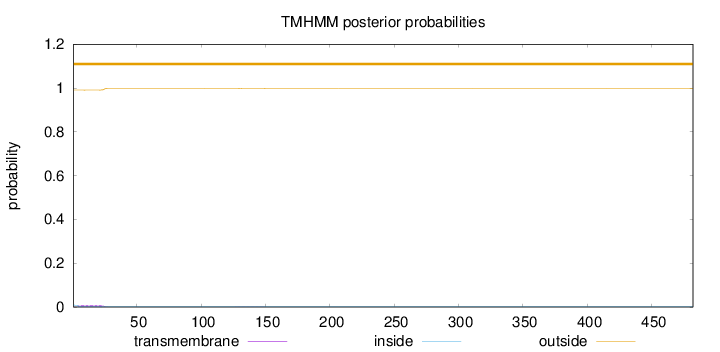
Length:
482
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19274
Exp number, first 60 AAs:
0.17515
Total prob of N-in:
0.00862
outside
1 - 482
Population Genetic Test Statistics
Pi
138.105854
Theta
170.974785
Tajima's D
-0.506269
CLR
3.09089
CSRT
0.24348782560872
Interpretation
Uncertain
