Gene
KWMTBOMO14318
Pre Gene Modal
BGIBMGA001235
Annotation
GTP_cyclohydrolase_I_isoform_B_[Bombyx_mori]
Full name
GTP cyclohydrolase 1
Alternative Name
GTP cyclohydrolase I
Protein punch
Protein punch
Location in the cell
Cytoplasmic Reliability : 1.773 Mitochondrial Reliability : 1.755
Sequence
CDS
ATGGCCGCCGTTAACGGAAAAGATTTGAACTCGGAAGCGGCAGTGCCGCCATCTTCGCTGATACGTCGCGTCAGTTCGCGATCCATACGTAATTCGATATCCGAGGAGAAAGAGAACGGTGACGCTCCTCAGAAGGGCTCGTTCGTTGGTAAATACAAAAAAGCTCCAGAAGCCCTGAAGAGCTTTAACTTGGCCGATGCGGAAAGTGAACTTGAAGTGCCCGGAACGCCAATGACGCCGAGGACTTCGACCACGCCTGGTCACGAGAACTGCACCTTCCACCATGACCTGGAACTGGACCACAGACCGCCCACCAGGGAGGCGTTGTTACCGGACATGGCTAACTCATACAGGCTTCTACTCACCGGCCTGGGGGAGGACCCTGAAAGGGCAGGGCTCTTGAAGACGCCCGAACGTGCCGCCAAAGCTATGCTCTTCTTCACGAAAGGATACGACCAAAGCTTGGAAGAGGTTCTGAACAACGCCATCTTCGATGAAGACACGGACGAGATGGTGGTCGTTAAGGATATCGAGATGTTTTCAATGTGCGAACATCATCTGGTGCCGTTTTACGGAAAGGTCTCCATCGGGTACCTGCCACAAGGAAAAATTCTGGGTCTCAGCAAACTGGCCAGGATCGTGGAAATATTCTCCCGCCGTCTTCAGGTACAAGAACGTCTGACGAAGCAGATAGCCATCGCGGTGACACAGGCCGTACGCCCGGCCGGCGTGGCGGTCGTCATTGAAGGAGTTCACATGTGCATGGTGATGCGAGGAGTCCAGAAGATCAACAGTAAGACGGTGACGTCGACGATGCTGGGCGTCTTCCGCGACGACCCGAAGACCCGGGAGGAGTTCCTCAACCTCGTCCACTCTAAGTGA
Protein
MAAVNGKDLNSEAAVPPSSLIRRVSSRSIRNSISEEKENGDAPQKGSFVGKYKKAPEALKSFNLADAESELEVPGTPMTPRTSTTPGHENCTFHHDLELDHRPPTREALLPDMANSYRLLLTGLGEDPERAGLLKTPERAAKAMLFFTKGYDQSLEEVLNNAIFDEDTDEMVVVKDIEMFSMCEHHLVPFYGKVSIGYLPQGKILGLSKLARIVEIFSRRLQVQERLTKQIAIAVTQAVRPAGVAVVIEGVHMCMVMRGVQKINSKTVTSTMLGVFRDDPKTREEFLNLVHSK
Summary
Description
Isoform B is required for eye pigment production, Isoform C may be required for normal embryonic development and segment pattern formation.
Catalytic Activity
GTP + H2O = 7,8-dihydroneopterin 3'-triphosphate + formate + H(+)
Subunit
Toroid-shaped homodecamer, composed of two pentamers of five dimers.
Similarity
Belongs to the GTP cyclohydrolase I family.
Keywords
Allosteric enzyme
Alternative splicing
Complete proteome
GTP-binding
Hydrolase
Metal-binding
Nucleotide-binding
Reference proteome
Tetrahydrobiopterin biosynthesis
Zinc
Feature
chain GTP cyclohydrolase 1
splice variant In isoform B.
splice variant In isoform B.
Uniprot
B7XAK4
A0A2H1W8H6
A0A2W1BVN8
A0A2A4J1K8
Q2V0Q0
S4NXA4
+ More
A0A3S2NFR0 B7XAK3 Q2V0Q1 H9IVF5 A0A1L8DZK3 A0A2A4J2T3 C4WXZ2 A0A2H8TPE0 A0A034WCM0 A0A0A1XN58 A0A0K8TYB3 W8B4A3 A0A1B6I2H8 A0A212FFR9 U5EVK2 A0A0T6B9V2 A0A1Y1LIQ3 A0A1J1IAL9 D2CFW6 A0A0L0BUZ2 T1P848 A0A1I8N9Z7 A0A0P9BXL2 A0A195BVR9 A0A158N914 E0VJS1 A0A151IBH4 A0A1I8QDD9 A0A1W4UMH8 A0A1I8QDE6 B4MQN1 A0A0J9U7H6 A0A151WNR5 A0A0Q5VKT5 A0A336ML79 A0A0R1DX11 P48596 A0A0Q9WFC2 A0A1A9UH08 B4J6Q1 A0A1A9Y0P3 A0A1B0BNJ2 B5E1M2 A0A1A9ZWC8 A0A1W4WUL6 A0A0M4EFC0 A0A1Q3EW34 A0A1B0G7Q9 A0A1A9WIP8 A0A0J9RHY2 A0A0Q9XL40 A0A0Q5VLL2 A0A034WE62 A0A0K8WLM4 R4WSU4 A0A0R1DRB3 A0A0A1XE91 A0A151JA93 A0A1S4F159 Q17IQ8 A0A1Q3EVT8 A0A1Q3EVU9 A0A2M4AIB9 A0A182U658 A0A1Q3EW49 A7UU97 A0A2P8ZJN3 A0A182FFA7 A0A2M4CSB5 A0A2M4AHD4 A0A2M4CRM0 A0A182WC45 A0A182NJ51 Q7Q5H2 A0A1Y9HAK5 A0A182I422 A0A182LTC4 A0A1Q3FUU0 A0A2M4AR74 A0A023FCW8 A0A1W7R8E5 B0X6U3 A0A1Y9H9G6 A0A1Q3FUH8 A0A2M4CRU7 A0A226EU62 A0A182JDF1 A0A195FU39 A0A182X8Y9 A0A0J7L1F9 A0A182GXL6 A0A182V9I3 A0A182LL00 A0A2M4BSK0
A0A3S2NFR0 B7XAK3 Q2V0Q1 H9IVF5 A0A1L8DZK3 A0A2A4J2T3 C4WXZ2 A0A2H8TPE0 A0A034WCM0 A0A0A1XN58 A0A0K8TYB3 W8B4A3 A0A1B6I2H8 A0A212FFR9 U5EVK2 A0A0T6B9V2 A0A1Y1LIQ3 A0A1J1IAL9 D2CFW6 A0A0L0BUZ2 T1P848 A0A1I8N9Z7 A0A0P9BXL2 A0A195BVR9 A0A158N914 E0VJS1 A0A151IBH4 A0A1I8QDD9 A0A1W4UMH8 A0A1I8QDE6 B4MQN1 A0A0J9U7H6 A0A151WNR5 A0A0Q5VKT5 A0A336ML79 A0A0R1DX11 P48596 A0A0Q9WFC2 A0A1A9UH08 B4J6Q1 A0A1A9Y0P3 A0A1B0BNJ2 B5E1M2 A0A1A9ZWC8 A0A1W4WUL6 A0A0M4EFC0 A0A1Q3EW34 A0A1B0G7Q9 A0A1A9WIP8 A0A0J9RHY2 A0A0Q9XL40 A0A0Q5VLL2 A0A034WE62 A0A0K8WLM4 R4WSU4 A0A0R1DRB3 A0A0A1XE91 A0A151JA93 A0A1S4F159 Q17IQ8 A0A1Q3EVT8 A0A1Q3EVU9 A0A2M4AIB9 A0A182U658 A0A1Q3EW49 A7UU97 A0A2P8ZJN3 A0A182FFA7 A0A2M4CSB5 A0A2M4AHD4 A0A2M4CRM0 A0A182WC45 A0A182NJ51 Q7Q5H2 A0A1Y9HAK5 A0A182I422 A0A182LTC4 A0A1Q3FUU0 A0A2M4AR74 A0A023FCW8 A0A1W7R8E5 B0X6U3 A0A1Y9H9G6 A0A1Q3FUH8 A0A2M4CRU7 A0A226EU62 A0A182JDF1 A0A195FU39 A0A182X8Y9 A0A0J7L1F9 A0A182GXL6 A0A182V9I3 A0A182LL00 A0A2M4BSK0
EC Number
3.5.4.16
Pubmed
18854583
28756777
16360951
26354079
23622113
19121390
+ More
25348373 25830018 24495485 22118469 28004739 18362917 19820115 26108605 25315136 17994087 21347285 20566863 22936249 17550304 8262960 10731132 12537572 12537569 3080426 15632085 23691247 17510324 12364791 14747013 17210077 29403074 25474469 26483478 20966253
25348373 25830018 24495485 22118469 28004739 18362917 19820115 26108605 25315136 17994087 21347285 20566863 22936249 17550304 8262960 10731132 12537572 12537569 3080426 15632085 23691247 17510324 12364791 14747013 17210077 29403074 25474469 26483478 20966253
EMBL
AB439288
BAH11150.1
ODYU01007026
SOQ49405.1
KZ149980
PZC75823.1
+ More
NWSH01003616 PCG66047.1 AB220982 KQ459386 BAE66651.1 KPJ01199.1 GAIX01012242 JAA80318.1 RSAL01000055 RVE49934.1 AB439287 BAH11149.1 AB220981 BAE66650.1 BABH01001014 BABH01001015 GFDF01002186 JAV11898.1 PCG66046.1 ABLF02034660 AK342930 BAH72762.1 GFXV01004210 MBW16015.1 GAKP01006885 JAC52067.1 GBXI01001528 JAD12764.1 GDHF01033071 JAI19243.1 GAMC01013143 JAB93412.1 GECU01026641 JAS81065.1 AGBW02008773 OWR52579.1 GANO01001821 JAB58050.1 LJIG01002827 KRT84113.1 GEZM01054659 JAV73503.1 CVRI01000047 CRK97335.1 DS497670 EFA13034.1 JRES01001422 KNC23049.1 KA644752 AFP59381.1 CH902619 KPU76231.1 KQ976405 KYM91891.1 ADTU01009025 ADTU01009026 ADTU01009027 ADTU01009028 DS235226 EEB13627.1 KQ978095 KYM97014.1 CH963849 EDW74420.1 CM002911 KMY95460.1 KQ982907 KYQ49460.1 CH954179 KQS62256.1 UFQT01001613 SSX31162.1 CM000158 KRJ99769.1 U01118 U01119 AF159422 AY382619 AY382620 AY382622 AY382623 AY382624 AY382625 AY382626 AY382628 AE013599 AY051890 CH940648 KRF79723.1 CH916367 EDW00954.1 JXJN01017419 CM000071 EDY69524.1 CP012524 ALC40860.1 GFDL01015627 JAV19418.1 CCAG010016891 KMY95462.1 CH933808 KRG05015.1 KQS62257.1 GAKP01006884 JAC52068.1 GDHF01000236 JAI52078.1 AK417787 BAN21002.1 KRJ99767.1 GBXI01017147 GBXI01005015 GBXI01002966 JAC97144.1 JAD09277.1 JAD11326.1 KQ979317 KYN21954.1 CH477237 EAT46580.1 GFDL01015624 JAV19421.1 GFDL01015614 JAV19431.1 GGFK01007037 MBW40358.1 GFDL01015617 JAV19428.1 AAAB01008960 EDO63875.1 PYGN01000035 PSN56709.1 GGFL01003883 MBW68061.1 GGFK01006811 MBW40132.1 GGFL01003804 MBW67982.1 EAA11896.4 AXCN02000457 APCN01002270 AXCM01005878 GFDL01003832 JAV31213.1 GGFK01009952 MBW43273.1 GBBI01000193 JAC18519.1 GEHC01000238 JAV47407.1 DS232424 EDS41614.1 GFDL01003830 JAV31215.1 GGFL01003884 MBW68062.1 LNIX01000002 OXA60687.1 KQ981268 KYN43941.1 LBMM01001332 KMQ96496.1 JXUM01095715 JXUM01095716 JXUM01095717 KQ564213 KXJ72579.1 GGFJ01006898 MBW56039.1
NWSH01003616 PCG66047.1 AB220982 KQ459386 BAE66651.1 KPJ01199.1 GAIX01012242 JAA80318.1 RSAL01000055 RVE49934.1 AB439287 BAH11149.1 AB220981 BAE66650.1 BABH01001014 BABH01001015 GFDF01002186 JAV11898.1 PCG66046.1 ABLF02034660 AK342930 BAH72762.1 GFXV01004210 MBW16015.1 GAKP01006885 JAC52067.1 GBXI01001528 JAD12764.1 GDHF01033071 JAI19243.1 GAMC01013143 JAB93412.1 GECU01026641 JAS81065.1 AGBW02008773 OWR52579.1 GANO01001821 JAB58050.1 LJIG01002827 KRT84113.1 GEZM01054659 JAV73503.1 CVRI01000047 CRK97335.1 DS497670 EFA13034.1 JRES01001422 KNC23049.1 KA644752 AFP59381.1 CH902619 KPU76231.1 KQ976405 KYM91891.1 ADTU01009025 ADTU01009026 ADTU01009027 ADTU01009028 DS235226 EEB13627.1 KQ978095 KYM97014.1 CH963849 EDW74420.1 CM002911 KMY95460.1 KQ982907 KYQ49460.1 CH954179 KQS62256.1 UFQT01001613 SSX31162.1 CM000158 KRJ99769.1 U01118 U01119 AF159422 AY382619 AY382620 AY382622 AY382623 AY382624 AY382625 AY382626 AY382628 AE013599 AY051890 CH940648 KRF79723.1 CH916367 EDW00954.1 JXJN01017419 CM000071 EDY69524.1 CP012524 ALC40860.1 GFDL01015627 JAV19418.1 CCAG010016891 KMY95462.1 CH933808 KRG05015.1 KQS62257.1 GAKP01006884 JAC52068.1 GDHF01000236 JAI52078.1 AK417787 BAN21002.1 KRJ99767.1 GBXI01017147 GBXI01005015 GBXI01002966 JAC97144.1 JAD09277.1 JAD11326.1 KQ979317 KYN21954.1 CH477237 EAT46580.1 GFDL01015624 JAV19421.1 GFDL01015614 JAV19431.1 GGFK01007037 MBW40358.1 GFDL01015617 JAV19428.1 AAAB01008960 EDO63875.1 PYGN01000035 PSN56709.1 GGFL01003883 MBW68061.1 GGFK01006811 MBW40132.1 GGFL01003804 MBW67982.1 EAA11896.4 AXCN02000457 APCN01002270 AXCM01005878 GFDL01003832 JAV31213.1 GGFK01009952 MBW43273.1 GBBI01000193 JAC18519.1 GEHC01000238 JAV47407.1 DS232424 EDS41614.1 GFDL01003830 JAV31215.1 GGFL01003884 MBW68062.1 LNIX01000002 OXA60687.1 KQ981268 KYN43941.1 LBMM01001332 KMQ96496.1 JXUM01095715 JXUM01095716 JXUM01095717 KQ564213 KXJ72579.1 GGFJ01006898 MBW56039.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000005204
UP000007819
UP000007151
+ More
UP000183832 UP000007266 UP000037069 UP000095301 UP000007801 UP000078540 UP000005205 UP000009046 UP000078542 UP000095300 UP000192221 UP000007798 UP000075809 UP000008711 UP000002282 UP000000803 UP000008792 UP000078200 UP000001070 UP000092443 UP000092460 UP000001819 UP000092445 UP000192223 UP000092553 UP000092444 UP000091820 UP000009192 UP000078492 UP000008820 UP000075902 UP000007062 UP000245037 UP000069272 UP000075920 UP000075884 UP000075886 UP000075840 UP000075883 UP000002320 UP000198287 UP000075880 UP000078541 UP000076407 UP000036403 UP000069940 UP000249989 UP000075903 UP000075882
UP000183832 UP000007266 UP000037069 UP000095301 UP000007801 UP000078540 UP000005205 UP000009046 UP000078542 UP000095300 UP000192221 UP000007798 UP000075809 UP000008711 UP000002282 UP000000803 UP000008792 UP000078200 UP000001070 UP000092443 UP000092460 UP000001819 UP000092445 UP000192223 UP000092553 UP000092444 UP000091820 UP000009192 UP000078492 UP000008820 UP000075902 UP000007062 UP000245037 UP000069272 UP000075920 UP000075884 UP000075886 UP000075840 UP000075883 UP000002320 UP000198287 UP000075880 UP000078541 UP000076407 UP000036403 UP000069940 UP000249989 UP000075903 UP000075882
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
B7XAK4
A0A2H1W8H6
A0A2W1BVN8
A0A2A4J1K8
Q2V0Q0
S4NXA4
+ More
A0A3S2NFR0 B7XAK3 Q2V0Q1 H9IVF5 A0A1L8DZK3 A0A2A4J2T3 C4WXZ2 A0A2H8TPE0 A0A034WCM0 A0A0A1XN58 A0A0K8TYB3 W8B4A3 A0A1B6I2H8 A0A212FFR9 U5EVK2 A0A0T6B9V2 A0A1Y1LIQ3 A0A1J1IAL9 D2CFW6 A0A0L0BUZ2 T1P848 A0A1I8N9Z7 A0A0P9BXL2 A0A195BVR9 A0A158N914 E0VJS1 A0A151IBH4 A0A1I8QDD9 A0A1W4UMH8 A0A1I8QDE6 B4MQN1 A0A0J9U7H6 A0A151WNR5 A0A0Q5VKT5 A0A336ML79 A0A0R1DX11 P48596 A0A0Q9WFC2 A0A1A9UH08 B4J6Q1 A0A1A9Y0P3 A0A1B0BNJ2 B5E1M2 A0A1A9ZWC8 A0A1W4WUL6 A0A0M4EFC0 A0A1Q3EW34 A0A1B0G7Q9 A0A1A9WIP8 A0A0J9RHY2 A0A0Q9XL40 A0A0Q5VLL2 A0A034WE62 A0A0K8WLM4 R4WSU4 A0A0R1DRB3 A0A0A1XE91 A0A151JA93 A0A1S4F159 Q17IQ8 A0A1Q3EVT8 A0A1Q3EVU9 A0A2M4AIB9 A0A182U658 A0A1Q3EW49 A7UU97 A0A2P8ZJN3 A0A182FFA7 A0A2M4CSB5 A0A2M4AHD4 A0A2M4CRM0 A0A182WC45 A0A182NJ51 Q7Q5H2 A0A1Y9HAK5 A0A182I422 A0A182LTC4 A0A1Q3FUU0 A0A2M4AR74 A0A023FCW8 A0A1W7R8E5 B0X6U3 A0A1Y9H9G6 A0A1Q3FUH8 A0A2M4CRU7 A0A226EU62 A0A182JDF1 A0A195FU39 A0A182X8Y9 A0A0J7L1F9 A0A182GXL6 A0A182V9I3 A0A182LL00 A0A2M4BSK0
A0A3S2NFR0 B7XAK3 Q2V0Q1 H9IVF5 A0A1L8DZK3 A0A2A4J2T3 C4WXZ2 A0A2H8TPE0 A0A034WCM0 A0A0A1XN58 A0A0K8TYB3 W8B4A3 A0A1B6I2H8 A0A212FFR9 U5EVK2 A0A0T6B9V2 A0A1Y1LIQ3 A0A1J1IAL9 D2CFW6 A0A0L0BUZ2 T1P848 A0A1I8N9Z7 A0A0P9BXL2 A0A195BVR9 A0A158N914 E0VJS1 A0A151IBH4 A0A1I8QDD9 A0A1W4UMH8 A0A1I8QDE6 B4MQN1 A0A0J9U7H6 A0A151WNR5 A0A0Q5VKT5 A0A336ML79 A0A0R1DX11 P48596 A0A0Q9WFC2 A0A1A9UH08 B4J6Q1 A0A1A9Y0P3 A0A1B0BNJ2 B5E1M2 A0A1A9ZWC8 A0A1W4WUL6 A0A0M4EFC0 A0A1Q3EW34 A0A1B0G7Q9 A0A1A9WIP8 A0A0J9RHY2 A0A0Q9XL40 A0A0Q5VLL2 A0A034WE62 A0A0K8WLM4 R4WSU4 A0A0R1DRB3 A0A0A1XE91 A0A151JA93 A0A1S4F159 Q17IQ8 A0A1Q3EVT8 A0A1Q3EVU9 A0A2M4AIB9 A0A182U658 A0A1Q3EW49 A7UU97 A0A2P8ZJN3 A0A182FFA7 A0A2M4CSB5 A0A2M4AHD4 A0A2M4CRM0 A0A182WC45 A0A182NJ51 Q7Q5H2 A0A1Y9HAK5 A0A182I422 A0A182LTC4 A0A1Q3FUU0 A0A2M4AR74 A0A023FCW8 A0A1W7R8E5 B0X6U3 A0A1Y9H9G6 A0A1Q3FUH8 A0A2M4CRU7 A0A226EU62 A0A182JDF1 A0A195FU39 A0A182X8Y9 A0A0J7L1F9 A0A182GXL6 A0A182V9I3 A0A182LL00 A0A2M4BSK0
PDB
1FB1
E-value=3.82793e-78,
Score=740
Ontologies
PATHWAY
GO
PANTHER
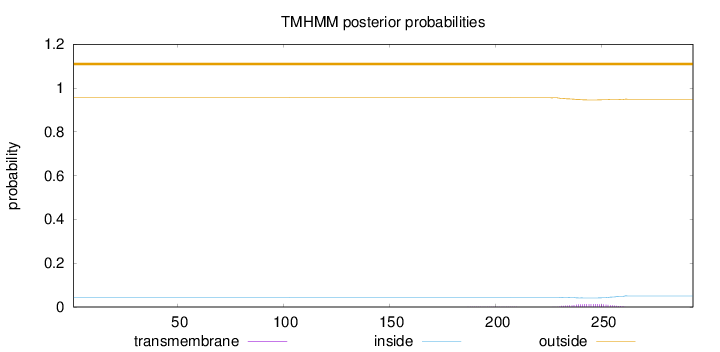
Topology
Length:
293
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28647
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04440
outside
1 - 293
Population Genetic Test Statistics
Pi
177.349104
Theta
154.815892
Tajima's D
0.26018
CLR
0.92308
CSRT
0.446127693615319
Interpretation
Uncertain
