Gene
KWMTBOMO14314 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008135
Annotation
PREDICTED:_uncharacterized_protein_LOC106130383_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.144 Extracellular Reliability : 1.528
Sequence
CDS
ATGCTTCAATTCGTCCTCCTACTTGTGGTCGCAGCTGCGGCATCCAAGAACTGCTCCTTCTATTTCGATGGTCATCCTTACTCAAGAACAGCCATATTATCCATTAAAGGCCTACCAACAAATTTGGTCTACAACCCTGCCAATCAAGATCTCCTATTCACTCTAATAGACCTGGAAACACTTCAAGACGACAACATCCAGACAAAAATGGACCAGTATATACTTCGAGAGGGGAATACGATCAAAATCGATAACGTTAGAGGTCAAGCTTCAGCGGTGGATATGAAACGAAACAAAGTGTACATAGCCAGCGATGACGGACTAAACGAACTAAACAGTACCGACAAAGCTAATTTCGTTGGTCTTAAAGATGACGATATCGTCCAGCTATACAAGCCGCGACACGGAGATGAATTGTACGCTGTTTTGTTTCCAGAAAACGCTGTTTACATAATCGACATAGAAAAAAACGAAAAGAGGAAAGTCGAATACATTCCCTGCGCTTTCTATTTGGGCGTAGATGACAAAGATAATATATTTTACGAATGTGATTCCAAATATGTTAAAGTTTTGCTCAAAGGCTTTCAAGAGCCCATTGAATTTGTTGGTATCGCTAAGAATTCCGGAAAAGCTATAGCTGTAGATGGAATTGGGAGAGTGATATTGGCTGCGACCGACGGTTTGTACCATCTGCGACCAGACAACGTGATCCCGAACAAACTGATGGAATTGGATTTCTATCCATCTGGTATAGCGTTTAATGAGGAAGACAATAGAATTTATTTATCGACGAATGATGTAATTTATAGATATAATCCATGTTTTCCTTTTATTTTGTGA
Protein
MLQFVLLLVVAAAASKNCSFYFDGHPYSRTAILSIKGLPTNLVYNPANQDLLFTLIDLETLQDDNIQTKMDQYILREGNTIKIDNVRGQASAVDMKRNKVYIASDDGLNELNSTDKANFVGLKDDDIVQLYKPRHGDELYAVLFPENAVYIIDIEKNEKRKVEYIPCAFYLGVDDKDNIFYECDSKYVKVLLKGFQEPIEFVGIAKNSGKAIAVDGIGRVILAATDGLYHLRPDNVIPNKLMELDFYPSGIAFNEEDNRIYLSTNDVIYRYNPCFPFIL
Summary
Uniprot
EMBL
BABH01026938
KZ149980
PZC75826.1
NWSH01003616
PCG66051.1
JTDY01000338
+ More
KOB77633.1 AK401563 KQ459386 BAM18185.1 KPJ01194.1 KQ460299 KPJ16155.1 ODYU01007026 SOQ49402.1 AGBW02010227 OWR48887.1 AK405267 BAM20629.1 PCG66052.1 RSAL01000055 RVE49936.1 PZC75827.1 KPJ01193.1 BABH01031160 KQ460254 KPJ16352.1 JTDY01003236 KOB69904.1 ODYU01010948 SOQ56376.1 NWSH01000545 PCG75751.1 NWSH01000044 PCG80297.1 KQ458859 KPJ04827.1 BABH01026891 KPJ01211.1 AGBW02013802 OWR42794.1 DMZT01000441 HAU81887.1
KOB77633.1 AK401563 KQ459386 BAM18185.1 KPJ01194.1 KQ460299 KPJ16155.1 ODYU01007026 SOQ49402.1 AGBW02010227 OWR48887.1 AK405267 BAM20629.1 PCG66052.1 RSAL01000055 RVE49936.1 PZC75827.1 KPJ01193.1 BABH01031160 KQ460254 KPJ16352.1 JTDY01003236 KOB69904.1 ODYU01010948 SOQ56376.1 NWSH01000545 PCG75751.1 NWSH01000044 PCG80297.1 KQ458859 KPJ04827.1 BABH01026891 KPJ01211.1 AGBW02013802 OWR42794.1 DMZT01000441 HAU81887.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
Ontologies
GO
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.963842
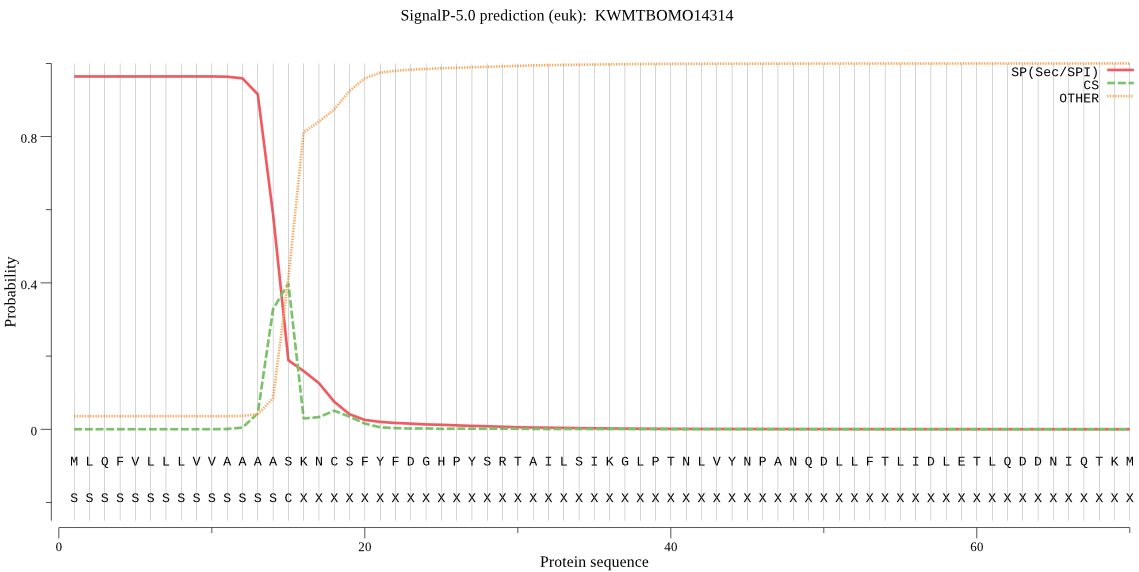
Length:
279
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17372
Exp number, first 60 AAs:
0.17151
Total prob of N-in:
0.00843
outside
1 - 279

Population Genetic Test Statistics
Pi
188.1518
Theta
140.46175
Tajima's D
0.911577
CLR
0
CSRT
0.631818409079546
Interpretation
Uncertain
