Gene
KWMTBOMO14309 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008137
Annotation
PREDICTED:_adrenodoxin-like_protein?_mitochondrial_[Amyelois_transitella]
Full name
Adrenodoxin-like protein, mitochondrial
Location in the cell
Cytoplasmic Reliability : 1.186 Mitochondrial Reliability : 1.546 Nuclear Reliability : 1.676
Sequence
CDS
ATGCTCCAATTATCAATTAAACGACTTATCTCGTGTAATAGACTGAGTGATTTGCGACGATCGTTTATTGTACAAAACAAGAGAATTCACACTACGAATCATTTAAAACACGGAGAATACGAATGGCAAGATCCGAAGTCTGAAGATGAAGTTGTGAACGTTGTATATGTTGATAAAGATGGAGTTAAAATAAATGTTAAAGGGAAAGTTGGTGATAACATTTTATATTTAGCACATAGATATGAGATTCCTATGGAAGGTGCATGTGAAGCTTCATTGGCGTGCACAACTTGCCACGTATACATTCCTCACGATTACTTAGATAAGCTCCCGCCATCGGAAGAGAAAGAAGATGATCTGTTAGATATGGCTCCGTTTCTCAAAGAAAATTCTAGATTAGGTTGTCAAATTGTTTTAACAAAAGAATTGGAAGGTATGGTAATACAATTACCGAAAGCAACTAGGAACTTTTACGTGGACGGACATAAACCTAAGCCACATTAA
Protein
MLQLSIKRLISCNRLSDLRRSFIVQNKRIHTTNHLKHGEYEWQDPKSEDEVVNVVYVDKDGVKINVKGKVGDNILYLAHRYEIPMEGACEASLACTTCHVYIPHDYLDKLPPSEEKEDDLLDMAPFLKENSRLGCQIVLTKELEGMVIQLPKATRNFYVDGHKPKPH
Summary
Cofactor
[2Fe-2S] cluster
Similarity
Belongs to the adrenodoxin/putidaredoxin family.
Keywords
2Fe-2S
Complete proteome
Electron transport
Iron
Iron-sulfur
Metal-binding
Mitochondrion
Reference proteome
Transit peptide
Transport
Feature
chain Adrenodoxin-like protein, mitochondrial
Uniprot
A0A2A4JUI6
H9JF40
A0A2H1WER5
A0A194Q6P2
A0A2W1BNI3
A0A2A4JSS4
+ More
A0A1I8MS10 A0A1S4EBK1 A0A0T6AYV6 K7IZ41 A0A023EHS9 A0A1Y1KC32 A0A1A9UGD8 A0A1A9ZU87 Q16WH1 A0A182QMT9 A0A1Q3F322 A0A1B0G3A7 A0A1A9W4C0 A0A1S4FDJ7 B0WC69 A0A212F580 A0A3L8DMV1 A0A182Y895 A0A0L0CPV7 A0A1I8NUX5 A0A0M9A1K7 A0A034WUP3 A0A182WI39 A0A1J1ISP8 A0A182R6P1 A0A0K8TWN1 A0A182IMD5 A0A0K8TLQ9 V5I6W7 A0A182UT85 Q7PTB2 A0A182I6C9 B4IYH2 W8BX71 A0A1B0F066 B3M6E5 N6T136 A0A1B0AVT2 A0A087ZX71 A0A182L411 A0A067QSH2 A0A182X268 B4LE23 A0A2A3EBU8 A0A3B0JPT1 B4KX22 A0A2H8TK74 A0A0M4F143 A0A2M4APU1 A0A182FFJ0 M9MRX8 P37193 B4HKN6 B4QMS4 H9XQB2 W5JCE0 B3NCE4 A0A182TSV1 C4WW10 B4PEY7 A0A2S2NBH5 A0A182K4C2 A0A1L8E101 A0A1L8E0H3 A0A1W4W7M1 A0A0P4VPS4 T1IDL6 B4MN40 A0A154PDV3 A0A336K2I9 G3MRS1 F2U9U9 E0W240 A0A026VUT7 A0A131YTG9 A0A3S1B9T6 D6W9A8 A0A182MST1 A0A3M7P1M7 E3NLA4 A0A260ZMV4 Q29F91 A0A310SRD9 A0A1D2N8U7 A0A1W4VB77 A0A210PMH6 A0A260Z821 A0A1S3H5L1 A0A261C683 B4H1M8 A0A2R7VW47 R7U2L3 A0A0C9S0I5
A0A1I8MS10 A0A1S4EBK1 A0A0T6AYV6 K7IZ41 A0A023EHS9 A0A1Y1KC32 A0A1A9UGD8 A0A1A9ZU87 Q16WH1 A0A182QMT9 A0A1Q3F322 A0A1B0G3A7 A0A1A9W4C0 A0A1S4FDJ7 B0WC69 A0A212F580 A0A3L8DMV1 A0A182Y895 A0A0L0CPV7 A0A1I8NUX5 A0A0M9A1K7 A0A034WUP3 A0A182WI39 A0A1J1ISP8 A0A182R6P1 A0A0K8TWN1 A0A182IMD5 A0A0K8TLQ9 V5I6W7 A0A182UT85 Q7PTB2 A0A182I6C9 B4IYH2 W8BX71 A0A1B0F066 B3M6E5 N6T136 A0A1B0AVT2 A0A087ZX71 A0A182L411 A0A067QSH2 A0A182X268 B4LE23 A0A2A3EBU8 A0A3B0JPT1 B4KX22 A0A2H8TK74 A0A0M4F143 A0A2M4APU1 A0A182FFJ0 M9MRX8 P37193 B4HKN6 B4QMS4 H9XQB2 W5JCE0 B3NCE4 A0A182TSV1 C4WW10 B4PEY7 A0A2S2NBH5 A0A182K4C2 A0A1L8E101 A0A1L8E0H3 A0A1W4W7M1 A0A0P4VPS4 T1IDL6 B4MN40 A0A154PDV3 A0A336K2I9 G3MRS1 F2U9U9 E0W240 A0A026VUT7 A0A131YTG9 A0A3S1B9T6 D6W9A8 A0A182MST1 A0A3M7P1M7 E3NLA4 A0A260ZMV4 Q29F91 A0A310SRD9 A0A1D2N8U7 A0A1W4VB77 A0A210PMH6 A0A260Z821 A0A1S3H5L1 A0A261C683 B4H1M8 A0A2R7VW47 R7U2L3 A0A0C9S0I5
Pubmed
19121390
26354079
28756777
25315136
20075255
24945155
+ More
26483478 28004739 17510324 22118469 30249741 25244985 26108605 25348373 26369729 12364791 14747013 17210077 17994087 24495485 23537049 20966253 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 3123699 12537569 22936249 20920257 23761445 17550304 27129103 22216098 20566863 24508170 26830274 18362917 19820115 30375419 15632085 27289101 28812685 23254933 26131772
26483478 28004739 17510324 22118469 30249741 25244985 26108605 25348373 26369729 12364791 14747013 17210077 17994087 24495485 23537049 20966253 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 3123699 12537569 22936249 20920257 23761445 17550304 27129103 22216098 20566863 24508170 26830274 18362917 19820115 30375419 15632085 27289101 28812685 23254933 26131772
EMBL
NWSH01000645
PCG75062.1
BABH01026930
ODYU01008180
SOQ51563.1
KQ459386
+ More
KPJ01203.1 KZ149980 PZC75831.1 PCG75061.1 LJIG01022499 KRT80250.1 JXUM01059303 GAPW01005189 KQ562045 JAC08409.1 KXJ76825.1 GEZM01093074 JAV56357.1 CH477566 EAT38926.1 AXCN02000573 GFDL01013118 JAV21927.1 CCAG010013743 DS231886 EDS43314.1 AGBW02010227 OWR48883.1 QOIP01000006 RLU21553.1 JRES01000080 KNC34380.1 KQ435794 KOX74033.1 GAKP01000895 JAC58057.1 CVRI01000059 CRL03245.1 GDHF01033452 JAI18862.1 GDAI01002324 JAI15279.1 GALX01007465 JAB61001.1 AAAB01008807 EAA04179.4 APCN01003611 CH916366 EDV97645.1 GAMC01000650 JAC05906.1 AJVK01017314 CH902618 EDV40794.1 APGK01048489 KB741112 KB632184 ENN73809.1 ERL89639.1 JXJN01004388 KK853004 KDR12591.1 CH940647 EDW70066.1 KZ288309 PBC28679.1 OUUW01000002 SPP77480.1 CH933809 EDW19665.1 GFXV01002721 MBW14526.1 CP012525 ALC44987.1 GGFK01009468 MBW42789.1 AE014296 ADV37512.1 X06542 AY113620 CH480815 EDW40839.1 CM000363 CM002912 EDX09828.1 KMY98609.1 BT133357 AFH01423.1 ADMH02001906 ETN60555.1 CH954178 EDV51174.1 ABLF02035500 AK341780 BAH72080.1 CM000159 EDW93042.1 GGMR01001507 MBY14126.1 GFDF01001909 JAV12175.1 GFDF01001910 JAV12174.1 GDKW01002805 JAI53790.1 ACPB03020268 CH963847 EDW73596.2 KQ434879 KZC09987.1 UFQS01000054 UFQT01000054 SSW98677.1 SSX19063.1 JO844572 AEO36189.1 GL832965 EGD73126.1 DS235874 EEB19696.1 KK107899 EZA47276.1 GEDV01006038 JAP82519.1 RQTK01000735 RUS75508.1 KQ971312 EEZ98188.2 AXCM01002970 REGN01014127 RMZ93012.1 DS268879 EFP04473.1 NMWX01000086 OZF86922.1 CH379067 EAL31364.1 KQ761323 OAD57891.1 LJIJ01000143 ODN01678.1 NEDP02005584 OWF37677.1 NIPN01000649 OZF81831.1 NIPN01000031 OZG18024.1 CH479202 EDW30205.1 KK854125 PTY11724.1 AMQN01010705 KB308442 ELT97886.1 GBZX01002611 JAG90129.1
KPJ01203.1 KZ149980 PZC75831.1 PCG75061.1 LJIG01022499 KRT80250.1 JXUM01059303 GAPW01005189 KQ562045 JAC08409.1 KXJ76825.1 GEZM01093074 JAV56357.1 CH477566 EAT38926.1 AXCN02000573 GFDL01013118 JAV21927.1 CCAG010013743 DS231886 EDS43314.1 AGBW02010227 OWR48883.1 QOIP01000006 RLU21553.1 JRES01000080 KNC34380.1 KQ435794 KOX74033.1 GAKP01000895 JAC58057.1 CVRI01000059 CRL03245.1 GDHF01033452 JAI18862.1 GDAI01002324 JAI15279.1 GALX01007465 JAB61001.1 AAAB01008807 EAA04179.4 APCN01003611 CH916366 EDV97645.1 GAMC01000650 JAC05906.1 AJVK01017314 CH902618 EDV40794.1 APGK01048489 KB741112 KB632184 ENN73809.1 ERL89639.1 JXJN01004388 KK853004 KDR12591.1 CH940647 EDW70066.1 KZ288309 PBC28679.1 OUUW01000002 SPP77480.1 CH933809 EDW19665.1 GFXV01002721 MBW14526.1 CP012525 ALC44987.1 GGFK01009468 MBW42789.1 AE014296 ADV37512.1 X06542 AY113620 CH480815 EDW40839.1 CM000363 CM002912 EDX09828.1 KMY98609.1 BT133357 AFH01423.1 ADMH02001906 ETN60555.1 CH954178 EDV51174.1 ABLF02035500 AK341780 BAH72080.1 CM000159 EDW93042.1 GGMR01001507 MBY14126.1 GFDF01001909 JAV12175.1 GFDF01001910 JAV12174.1 GDKW01002805 JAI53790.1 ACPB03020268 CH963847 EDW73596.2 KQ434879 KZC09987.1 UFQS01000054 UFQT01000054 SSW98677.1 SSX19063.1 JO844572 AEO36189.1 GL832965 EGD73126.1 DS235874 EEB19696.1 KK107899 EZA47276.1 GEDV01006038 JAP82519.1 RQTK01000735 RUS75508.1 KQ971312 EEZ98188.2 AXCM01002970 REGN01014127 RMZ93012.1 DS268879 EFP04473.1 NMWX01000086 OZF86922.1 CH379067 EAL31364.1 KQ761323 OAD57891.1 LJIJ01000143 ODN01678.1 NEDP02005584 OWF37677.1 NIPN01000649 OZF81831.1 NIPN01000031 OZG18024.1 CH479202 EDW30205.1 KK854125 PTY11724.1 AMQN01010705 KB308442 ELT97886.1 GBZX01002611 JAG90129.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000095301
UP000079169
UP000002358
+ More
UP000069940 UP000249989 UP000078200 UP000092445 UP000008820 UP000075886 UP000092444 UP000091820 UP000002320 UP000007151 UP000279307 UP000076408 UP000037069 UP000095300 UP000053105 UP000075920 UP000183832 UP000075900 UP000075880 UP000075903 UP000007062 UP000075840 UP000001070 UP000092462 UP000007801 UP000019118 UP000030742 UP000092460 UP000005203 UP000075882 UP000027135 UP000076407 UP000008792 UP000242457 UP000268350 UP000009192 UP000092553 UP000069272 UP000000803 UP000001292 UP000000304 UP000000673 UP000008711 UP000075902 UP000007819 UP000002282 UP000075881 UP000192223 UP000015103 UP000007798 UP000076502 UP000007799 UP000009046 UP000053097 UP000271974 UP000007266 UP000075883 UP000276133 UP000008281 UP000216624 UP000001819 UP000094527 UP000192221 UP000242188 UP000216463 UP000085678 UP000008744 UP000014760
UP000069940 UP000249989 UP000078200 UP000092445 UP000008820 UP000075886 UP000092444 UP000091820 UP000002320 UP000007151 UP000279307 UP000076408 UP000037069 UP000095300 UP000053105 UP000075920 UP000183832 UP000075900 UP000075880 UP000075903 UP000007062 UP000075840 UP000001070 UP000092462 UP000007801 UP000019118 UP000030742 UP000092460 UP000005203 UP000075882 UP000027135 UP000076407 UP000008792 UP000242457 UP000268350 UP000009192 UP000092553 UP000069272 UP000000803 UP000001292 UP000000304 UP000000673 UP000008711 UP000075902 UP000007819 UP000002282 UP000075881 UP000192223 UP000015103 UP000007798 UP000076502 UP000007799 UP000009046 UP000053097 UP000271974 UP000007266 UP000075883 UP000276133 UP000008281 UP000216624 UP000001819 UP000094527 UP000192221 UP000242188 UP000216463 UP000085678 UP000008744 UP000014760
Pfam
PF00111 Fer2
Interpro
SUPFAM
SSF54292
SSF54292
Gene 3D
CDD
ProteinModelPortal
A0A2A4JUI6
H9JF40
A0A2H1WER5
A0A194Q6P2
A0A2W1BNI3
A0A2A4JSS4
+ More
A0A1I8MS10 A0A1S4EBK1 A0A0T6AYV6 K7IZ41 A0A023EHS9 A0A1Y1KC32 A0A1A9UGD8 A0A1A9ZU87 Q16WH1 A0A182QMT9 A0A1Q3F322 A0A1B0G3A7 A0A1A9W4C0 A0A1S4FDJ7 B0WC69 A0A212F580 A0A3L8DMV1 A0A182Y895 A0A0L0CPV7 A0A1I8NUX5 A0A0M9A1K7 A0A034WUP3 A0A182WI39 A0A1J1ISP8 A0A182R6P1 A0A0K8TWN1 A0A182IMD5 A0A0K8TLQ9 V5I6W7 A0A182UT85 Q7PTB2 A0A182I6C9 B4IYH2 W8BX71 A0A1B0F066 B3M6E5 N6T136 A0A1B0AVT2 A0A087ZX71 A0A182L411 A0A067QSH2 A0A182X268 B4LE23 A0A2A3EBU8 A0A3B0JPT1 B4KX22 A0A2H8TK74 A0A0M4F143 A0A2M4APU1 A0A182FFJ0 M9MRX8 P37193 B4HKN6 B4QMS4 H9XQB2 W5JCE0 B3NCE4 A0A182TSV1 C4WW10 B4PEY7 A0A2S2NBH5 A0A182K4C2 A0A1L8E101 A0A1L8E0H3 A0A1W4W7M1 A0A0P4VPS4 T1IDL6 B4MN40 A0A154PDV3 A0A336K2I9 G3MRS1 F2U9U9 E0W240 A0A026VUT7 A0A131YTG9 A0A3S1B9T6 D6W9A8 A0A182MST1 A0A3M7P1M7 E3NLA4 A0A260ZMV4 Q29F91 A0A310SRD9 A0A1D2N8U7 A0A1W4VB77 A0A210PMH6 A0A260Z821 A0A1S3H5L1 A0A261C683 B4H1M8 A0A2R7VW47 R7U2L3 A0A0C9S0I5
A0A1I8MS10 A0A1S4EBK1 A0A0T6AYV6 K7IZ41 A0A023EHS9 A0A1Y1KC32 A0A1A9UGD8 A0A1A9ZU87 Q16WH1 A0A182QMT9 A0A1Q3F322 A0A1B0G3A7 A0A1A9W4C0 A0A1S4FDJ7 B0WC69 A0A212F580 A0A3L8DMV1 A0A182Y895 A0A0L0CPV7 A0A1I8NUX5 A0A0M9A1K7 A0A034WUP3 A0A182WI39 A0A1J1ISP8 A0A182R6P1 A0A0K8TWN1 A0A182IMD5 A0A0K8TLQ9 V5I6W7 A0A182UT85 Q7PTB2 A0A182I6C9 B4IYH2 W8BX71 A0A1B0F066 B3M6E5 N6T136 A0A1B0AVT2 A0A087ZX71 A0A182L411 A0A067QSH2 A0A182X268 B4LE23 A0A2A3EBU8 A0A3B0JPT1 B4KX22 A0A2H8TK74 A0A0M4F143 A0A2M4APU1 A0A182FFJ0 M9MRX8 P37193 B4HKN6 B4QMS4 H9XQB2 W5JCE0 B3NCE4 A0A182TSV1 C4WW10 B4PEY7 A0A2S2NBH5 A0A182K4C2 A0A1L8E101 A0A1L8E0H3 A0A1W4W7M1 A0A0P4VPS4 T1IDL6 B4MN40 A0A154PDV3 A0A336K2I9 G3MRS1 F2U9U9 E0W240 A0A026VUT7 A0A131YTG9 A0A3S1B9T6 D6W9A8 A0A182MST1 A0A3M7P1M7 E3NLA4 A0A260ZMV4 Q29F91 A0A310SRD9 A0A1D2N8U7 A0A1W4VB77 A0A210PMH6 A0A260Z821 A0A1S3H5L1 A0A261C683 B4H1M8 A0A2R7VW47 R7U2L3 A0A0C9S0I5
PDB
2Y5C
E-value=1.35883e-33,
Score=352
Ontologies
KEGG
GO
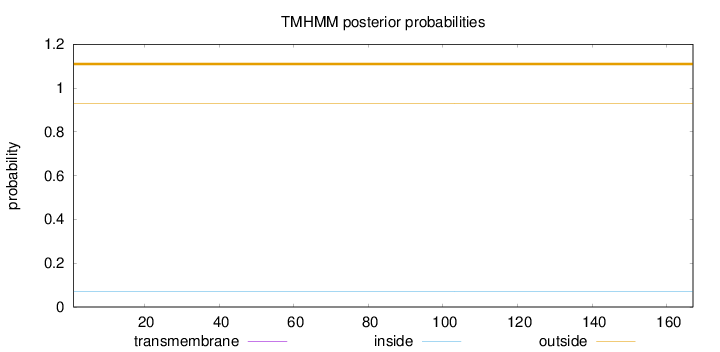
Topology
Subcellular location
Mitochondrion matrix
Length:
167
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00041
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06922
outside
1 - 167
Population Genetic Test Statistics
Pi
326.550684
Theta
185.127357
Tajima's D
2.41561
CLR
0.126788
CSRT
0.938403079846008
Interpretation
Uncertain
