Gene
KWMTBOMO14302
Pre Gene Modal
BGIBMGA008181
Annotation
PREDICTED:_protein-glutamate_O-methyltransferase-like_[Bombyx_mori]
Full name
Protein-glutamate O-methyltransferase
Alternative Name
Acidic residue methyltransferase 1
Location in the cell
Cytoplasmic Reliability : 2.251 Extracellular Reliability : 1.693
Sequence
CDS
ATGTTAATTTTCAGCAAAACTCTCAAGAATTTCGATCCGTTCGCTGACCAAAAGCAGAAATCGTTCGAAGGAAGCTTGGAGCTGATGTGTATTGTGGCCGATAAGCTAGTTAATATGTTATCTAAGAGCGATAAAGATAAATGTAAAAGTGATTTTGTTACTTTGCTGAAGCTCTGTCTGTGGGCCAACAAATGCGATTTGTCGTTGTCCAGTGGCGAGCAAGTGTTTTTGAACGGGAACAATGCTGACGGAAAAGCTACGACATCAAACATCATAGATCCATTCCAAATGATTCTCGATTTTAAGGATAAGTTATTAGTTGATGATACGTCAAAAATAGTCGATCAGGTGACGAATAAAGCTGAAGCCATGATGAAGAACTTAGAGGCGCCGGCTTCGGCTGCGGACGGCGCCAAAGGCTCTCCGCCCGAGACGCCCGCTGCTGAAGCCGCTGATGTCCCTTGTCCGACAAAGATGACATCACCTCAGCACGTCATGTTCGATATAGTATGCGATAATGCTGGGTACGAGTTATTCGCCGATTTGTGTTTAGCACATTTTCTGATATCACAGGGGATCGTGCAAAAGATTCGGTTCCACGTGAAGAACATACCTTGGTTTGTGTCTGACGTCACGCCCGTAGATTTTAAGTGGGTGATCGACTCATGCCTCAAAGCCAGCTACAGGAGAGAAGTCCCCCAGCAGCCGACAGAAGCGGCCGAGGGCGAGGTCCCCCCCGAGCCGGAGCCCCCCCGGGTGATAGGCGCGGACAATCTGCGCGCCCTGGCGGAGCAGTGGACCAAATACGTAGACGAAGGCACATTTGTAGTATTGCACGACGACTTCTGGACCTCCCCCCACGTGTACAGGGACATGAAAAGGCACGCGCCCTCTCTGTACCGCCAGCTGCAGTACGCGGTCGCGCTGCTGTTTAAGGGCGACCTCAACTACAGGAAGCTGCTCTGTGAGAAGAACGTGCCACCCACCAGTGGGCTGGAGGCCGCCTTGCAGGGCTTCCTCCCGGCTCCCGTGATCGCGGTGCGCACGGTGAAGGCCGACCTGATATGCGGGCTGCCGAAGGGTAAGTGGGACCAGCTGAACGCGCTCGACGACAAGTGGATGCAGAAGGGCGACTTCGGCGTGATTCAGTATTGCGGTAAAGTGGAACCACTAAAGGTAGCTGACCGGCCATGCGATCAGTACGGAGACGTCTGCAGGGGAGCCGGCTGTGGTCCACGGACCGATATGTGA
Protein
MLIFSKTLKNFDPFADQKQKSFEGSLELMCIVADKLVNMLSKSDKDKCKSDFVTLLKLCLWANKCDLSLSSGEQVFLNGNNADGKATTSNIIDPFQMILDFKDKLLVDDTSKIVDQVTNKAEAMMKNLEAPASAADGAKGSPPETPAAEAADVPCPTKMTSPQHVMFDIVCDNAGYELFADLCLAHFLISQGIVQKIRFHVKNIPWFVSDVTPVDFKWVIDSCLKASYRREVPQQPTEAAEGEVPPEPEPPRVIGADNLRALAEQWTKYVDEGTFVVLHDDFWTSPHVYRDMKRHAPSLYRQLQYAVALLFKGDLNYRKLLCEKNVPPTSGLEAALQGFLPAPVIAVRTVKADLICGLPKGKWDQLNALDDKWMQKGDFGVIQYCGKVEPLKVADRPCDQYGDVCRGAGCGPRTDM
Summary
Description
O-methyltransferase that methylates glutamate residues of target proteins to form gamma-glutamyl methyl ester residues.
O-methyltransferase that methylates glutamate residues of target proteins to form gamma-glutamyl methyl ester residues. Methylates PCNA, suggesting it is involved in the DNA damage response.
O-methyltransferase that methylates glutamate residues of target proteins to form gamma-glutamyl methyl ester residues. Methylates PCNA, suggesting it is involved in the DNA damage response.
Catalytic Activity
L-glutamyl-[protein] + S-adenosyl-L-methionine = [protein]-L-glutamate 5-O-methyl ester + S-adenosyl-L-homocysteine
Similarity
Belongs to the ARMT1 family.
Belongs to the nuclear hormone receptor family.
Belongs to the nuclear hormone receptor family.
Keywords
Complete proteome
Methyltransferase
Reference proteome
S-adenosyl-L-methionine
Transferase
Acetylation
DNA damage
Phosphoprotein
Feature
chain Protein-glutamate O-methyltransferase
Uniprot
A0A194Q6Q1
A0A2H1WEG1
A0A2A4K8X4
A0A212EHH4
A0A1Y1NGM0
A0A1Y1NDN9
+ More
A0A2J7PH50 A0A2J7PH33 A0A2J7PH49 B4MS52 B2GSN3 Q58EM4 A0A2R8Q952 A0A2M4ASK7 A0A2M4ASX4 W5JL90 A0A067R5A0 A0A182JEN4 A0A0V1HZ66 A0A0V1N7F3 A0A182KV33 A0A182TI69 A0A182I5X9 A0A182FLH5 Q7QE00 A0A1W4WPV1 A0A182V962 A0A1S4H4R8 A0A182WXV8 A0A1S3KC65 A0A0V1FWZ3 A0A0V0XMT8 A0A0V0S019 W5NHJ5 A0A3N0XHY9 A0A0V0VPJ8 A0A182PRM2 W5NHJ0 A0A2P8XFV5 A0A1Q3FBQ4 A0A0L7QPU9 A0A0V0WVG3 A0A0V0WVH4 A0A3P8TIM9 W5NZ43 A3KMX8 A0A384BCB7 L8IS35 A0A212C7N2 A0A3B1K347 F1MC76 A0A384BBS2 A0A3Q7Q1E0 A0A341BPG1 T1JVG8 A0A341BN45 G1LX50 A0A369S2H7 D2HE39 A0A1J1I8Q8 A0A341BPB3 A0A340XFD1 F1L8R3 A0A3Q7NKQ2 A0A087ZRY9 M3WCV6 A0A2U3XVN9 A0A2Y9HBH9 A0A341BN37 B3SAB3 A0A0M5J4P6 A0A2U3W7E0 A0A3Q0E9V3 A0A1U7UD16 B4LND7 C0MIF3 A0AMV9 A0A2S2PNZ5 A0A0J9RI22 A0A0N5AQK0 A0AMW1 B4I7Z2 A0A3L7I625 A0A2J8XAI3 A0A0P6G942 G3GU16 A0A0P5W7R7 A0A0P6D7U4 A0A3Q0CI59 A0A1U7Q3K0
A0A2J7PH50 A0A2J7PH33 A0A2J7PH49 B4MS52 B2GSN3 Q58EM4 A0A2R8Q952 A0A2M4ASK7 A0A2M4ASX4 W5JL90 A0A067R5A0 A0A182JEN4 A0A0V1HZ66 A0A0V1N7F3 A0A182KV33 A0A182TI69 A0A182I5X9 A0A182FLH5 Q7QE00 A0A1W4WPV1 A0A182V962 A0A1S4H4R8 A0A182WXV8 A0A1S3KC65 A0A0V1FWZ3 A0A0V0XMT8 A0A0V0S019 W5NHJ5 A0A3N0XHY9 A0A0V0VPJ8 A0A182PRM2 W5NHJ0 A0A2P8XFV5 A0A1Q3FBQ4 A0A0L7QPU9 A0A0V0WVG3 A0A0V0WVH4 A0A3P8TIM9 W5NZ43 A3KMX8 A0A384BCB7 L8IS35 A0A212C7N2 A0A3B1K347 F1MC76 A0A384BBS2 A0A3Q7Q1E0 A0A341BPG1 T1JVG8 A0A341BN45 G1LX50 A0A369S2H7 D2HE39 A0A1J1I8Q8 A0A341BPB3 A0A340XFD1 F1L8R3 A0A3Q7NKQ2 A0A087ZRY9 M3WCV6 A0A2U3XVN9 A0A2Y9HBH9 A0A341BN37 B3SAB3 A0A0M5J4P6 A0A2U3W7E0 A0A3Q0E9V3 A0A1U7UD16 B4LND7 C0MIF3 A0AMV9 A0A2S2PNZ5 A0A0J9RI22 A0A0N5AQK0 A0AMW1 B4I7Z2 A0A3L7I625 A0A2J8XAI3 A0A0P6G942 G3GU16 A0A0P5W7R7 A0A0P6D7U4 A0A3Q0CI59 A0A1U7Q3K0
EC Number
2.1.1.-
Pubmed
EMBL
KQ459386
KPJ01213.1
ODYU01008121
SOQ51453.1
NWSH01000023
PCG80647.1
+ More
AGBW02014889 OWR40935.1 GEZM01005626 JAV96030.1 GEZM01005625 JAV96032.1 NEVH01025150 PNF15649.1 PNF15647.1 PNF15648.1 CH963850 EDW74941.1 BC165582 AAI65582.1 BC091842 CR926887 GGFK01010429 MBW43750.1 GGFK01010539 MBW43860.1 ADMH02000758 ETN65142.1 KK852954 KDR13276.1 AXCP01007877 JYDP01000014 KRZ15897.1 JYDO01000004 KRZ79953.1 APCN01003537 AAAB01008848 EAA07056.3 JYDT01000020 KRY90586.1 JYDU01000204 JYDR01000106 JYDS01000090 KRX89249.1 KRY68687.1 KRZ26077.1 JYDL01000052 KRX20104.1 AHAT01015749 AHAT01015750 AHAT01015751 RJVU01072567 ROI33809.1 JYDN01000017 KRX65439.1 PYGN01002299 PSN30855.1 GFDL01010039 JAV25006.1 KQ414799 KOC60655.1 JYDK01000049 KRX79755.1 KRX79756.1 AMGL01108377 BC133378 JH880723 ELR59385.1 MKHE01000026 OWK01991.1 CAEY01000794 ACTA01075821 NOWV01000107 RDD40253.1 GL192740 EFB19021.1 CVRI01000043 CRK95956.1 JI173974 ADY46517.1 AANG04000169 DS985261 EDV20231.1 CP012524 ALC41194.1 CH940648 EDW61089.1 FM245125 CAR93051.1 AM293997 CAL25899.1 GGMR01018524 MBY31143.1 CM002911 KMY95698.1 AM293999 CAL25901.1 CH480824 EDW56717.1 RAZU01000095 RLQ73593.1 NDHI03003369 PNJ79019.1 GDIQ01036871 JAN57866.1 JH000025 EGV95942.1 GDIQ01230049 GDIQ01190024 GDIP01090106 LRGB01003389 JAK21676.1 JAM13609.1 KZS02859.1 GDIQ01230050 GDIQ01082159 JAN12578.1
AGBW02014889 OWR40935.1 GEZM01005626 JAV96030.1 GEZM01005625 JAV96032.1 NEVH01025150 PNF15649.1 PNF15647.1 PNF15648.1 CH963850 EDW74941.1 BC165582 AAI65582.1 BC091842 CR926887 GGFK01010429 MBW43750.1 GGFK01010539 MBW43860.1 ADMH02000758 ETN65142.1 KK852954 KDR13276.1 AXCP01007877 JYDP01000014 KRZ15897.1 JYDO01000004 KRZ79953.1 APCN01003537 AAAB01008848 EAA07056.3 JYDT01000020 KRY90586.1 JYDU01000204 JYDR01000106 JYDS01000090 KRX89249.1 KRY68687.1 KRZ26077.1 JYDL01000052 KRX20104.1 AHAT01015749 AHAT01015750 AHAT01015751 RJVU01072567 ROI33809.1 JYDN01000017 KRX65439.1 PYGN01002299 PSN30855.1 GFDL01010039 JAV25006.1 KQ414799 KOC60655.1 JYDK01000049 KRX79755.1 KRX79756.1 AMGL01108377 BC133378 JH880723 ELR59385.1 MKHE01000026 OWK01991.1 CAEY01000794 ACTA01075821 NOWV01000107 RDD40253.1 GL192740 EFB19021.1 CVRI01000043 CRK95956.1 JI173974 ADY46517.1 AANG04000169 DS985261 EDV20231.1 CP012524 ALC41194.1 CH940648 EDW61089.1 FM245125 CAR93051.1 AM293997 CAL25899.1 GGMR01018524 MBY31143.1 CM002911 KMY95698.1 AM293999 CAL25901.1 CH480824 EDW56717.1 RAZU01000095 RLQ73593.1 NDHI03003369 PNJ79019.1 GDIQ01036871 JAN57866.1 JH000025 EGV95942.1 GDIQ01230049 GDIQ01190024 GDIP01090106 LRGB01003389 JAK21676.1 JAM13609.1 KZS02859.1 GDIQ01230050 GDIQ01082159 JAN12578.1
Proteomes
UP000053268
UP000218220
UP000007151
UP000235965
UP000007798
UP000000437
+ More
UP000000673 UP000027135 UP000075880 UP000055024 UP000054843 UP000075882 UP000075902 UP000075840 UP000069272 UP000007062 UP000192223 UP000075903 UP000076407 UP000085678 UP000054995 UP000054632 UP000054805 UP000054815 UP000054630 UP000018468 UP000054681 UP000075885 UP000245037 UP000053825 UP000054673 UP000265080 UP000002356 UP000009136 UP000261681 UP000018467 UP000286641 UP000252040 UP000015104 UP000008912 UP000253843 UP000183832 UP000265300 UP000005203 UP000011712 UP000245341 UP000248481 UP000009022 UP000092553 UP000245340 UP000189704 UP000008792 UP000046393 UP000001292 UP000273346 UP000001075 UP000076858 UP000189706
UP000000673 UP000027135 UP000075880 UP000055024 UP000054843 UP000075882 UP000075902 UP000075840 UP000069272 UP000007062 UP000192223 UP000075903 UP000076407 UP000085678 UP000054995 UP000054632 UP000054805 UP000054815 UP000054630 UP000018468 UP000054681 UP000075885 UP000245037 UP000053825 UP000054673 UP000265080 UP000002356 UP000009136 UP000261681 UP000018467 UP000286641 UP000252040 UP000015104 UP000008912 UP000253843 UP000183832 UP000265300 UP000005203 UP000011712 UP000245341 UP000248481 UP000009022 UP000092553 UP000245340 UP000189704 UP000008792 UP000046393 UP000001292 UP000273346 UP000001075 UP000076858 UP000189706
Pfam
Interpro
IPR039763
ARMT1
+ More
IPR036075 AF1104-like_sf
IPR002791 DUF89
IPR013087 Znf_C2H2_type
IPR002999 Tudor
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR035979 RBD_domain_sf
IPR001292 Oestr_rcpt
IPR035500 NHR-like_dom_sf
IPR001723 Nuclear_hrmn_rcpt
IPR013088 Znf_NHR/GATA
IPR000536 Nucl_hrmn_rcpt_lig-bd
IPR001628 Znf_hrmn_rcpt
IPR036075 AF1104-like_sf
IPR002791 DUF89
IPR013087 Znf_C2H2_type
IPR002999 Tudor
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR035979 RBD_domain_sf
IPR001292 Oestr_rcpt
IPR035500 NHR-like_dom_sf
IPR001723 Nuclear_hrmn_rcpt
IPR013088 Znf_NHR/GATA
IPR000536 Nucl_hrmn_rcpt_lig-bd
IPR001628 Znf_hrmn_rcpt
Gene 3D
ProteinModelPortal
A0A194Q6Q1
A0A2H1WEG1
A0A2A4K8X4
A0A212EHH4
A0A1Y1NGM0
A0A1Y1NDN9
+ More
A0A2J7PH50 A0A2J7PH33 A0A2J7PH49 B4MS52 B2GSN3 Q58EM4 A0A2R8Q952 A0A2M4ASK7 A0A2M4ASX4 W5JL90 A0A067R5A0 A0A182JEN4 A0A0V1HZ66 A0A0V1N7F3 A0A182KV33 A0A182TI69 A0A182I5X9 A0A182FLH5 Q7QE00 A0A1W4WPV1 A0A182V962 A0A1S4H4R8 A0A182WXV8 A0A1S3KC65 A0A0V1FWZ3 A0A0V0XMT8 A0A0V0S019 W5NHJ5 A0A3N0XHY9 A0A0V0VPJ8 A0A182PRM2 W5NHJ0 A0A2P8XFV5 A0A1Q3FBQ4 A0A0L7QPU9 A0A0V0WVG3 A0A0V0WVH4 A0A3P8TIM9 W5NZ43 A3KMX8 A0A384BCB7 L8IS35 A0A212C7N2 A0A3B1K347 F1MC76 A0A384BBS2 A0A3Q7Q1E0 A0A341BPG1 T1JVG8 A0A341BN45 G1LX50 A0A369S2H7 D2HE39 A0A1J1I8Q8 A0A341BPB3 A0A340XFD1 F1L8R3 A0A3Q7NKQ2 A0A087ZRY9 M3WCV6 A0A2U3XVN9 A0A2Y9HBH9 A0A341BN37 B3SAB3 A0A0M5J4P6 A0A2U3W7E0 A0A3Q0E9V3 A0A1U7UD16 B4LND7 C0MIF3 A0AMV9 A0A2S2PNZ5 A0A0J9RI22 A0A0N5AQK0 A0AMW1 B4I7Z2 A0A3L7I625 A0A2J8XAI3 A0A0P6G942 G3GU16 A0A0P5W7R7 A0A0P6D7U4 A0A3Q0CI59 A0A1U7Q3K0
A0A2J7PH50 A0A2J7PH33 A0A2J7PH49 B4MS52 B2GSN3 Q58EM4 A0A2R8Q952 A0A2M4ASK7 A0A2M4ASX4 W5JL90 A0A067R5A0 A0A182JEN4 A0A0V1HZ66 A0A0V1N7F3 A0A182KV33 A0A182TI69 A0A182I5X9 A0A182FLH5 Q7QE00 A0A1W4WPV1 A0A182V962 A0A1S4H4R8 A0A182WXV8 A0A1S3KC65 A0A0V1FWZ3 A0A0V0XMT8 A0A0V0S019 W5NHJ5 A0A3N0XHY9 A0A0V0VPJ8 A0A182PRM2 W5NHJ0 A0A2P8XFV5 A0A1Q3FBQ4 A0A0L7QPU9 A0A0V0WVG3 A0A0V0WVH4 A0A3P8TIM9 W5NZ43 A3KMX8 A0A384BCB7 L8IS35 A0A212C7N2 A0A3B1K347 F1MC76 A0A384BBS2 A0A3Q7Q1E0 A0A341BPG1 T1JVG8 A0A341BN45 G1LX50 A0A369S2H7 D2HE39 A0A1J1I8Q8 A0A341BPB3 A0A340XFD1 F1L8R3 A0A3Q7NKQ2 A0A087ZRY9 M3WCV6 A0A2U3XVN9 A0A2Y9HBH9 A0A341BN37 B3SAB3 A0A0M5J4P6 A0A2U3W7E0 A0A3Q0E9V3 A0A1U7UD16 B4LND7 C0MIF3 A0AMV9 A0A2S2PNZ5 A0A0J9RI22 A0A0N5AQK0 A0AMW1 B4I7Z2 A0A3L7I625 A0A2J8XAI3 A0A0P6G942 G3GU16 A0A0P5W7R7 A0A0P6D7U4 A0A3Q0CI59 A0A1U7Q3K0
PDB
5BY0
E-value=1.68722e-23,
Score=271
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
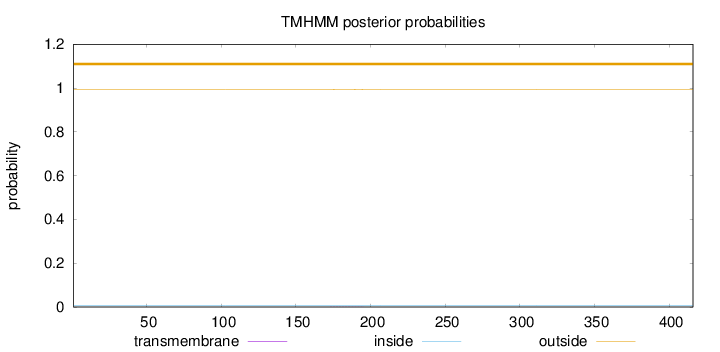
Length:
416
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0152
Exp number, first 60 AAs:
0.00034
Total prob of N-in:
0.00595
outside
1 - 416
Population Genetic Test Statistics
Pi
251.46744
Theta
200.68686
Tajima's D
1.518747
CLR
0.090235
CSRT
0.791760411979401
Interpretation
Uncertain
