Gene
KWMTBOMO14298
Pre Gene Modal
BGIBMGA008180
Annotation
Ommochrome-binding_protein_precursor_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.46
Sequence
CDS
ATGCTCATGAAATATTTAGTAATGTTAATTGTCGCGAGTGCAGAAGCGAAAATACGAATCTATCTGACTGATACACCGCTACAGATAATAGGGACGAGATTGTACAAGAAAGAACTACTAACAGACCTATACCAGGCGCCAAAAGAATTGACATATGATTCGTCGAGTAGAAACCTTTATTTCATGTACATGGATGACTCTCTACAGAATTCGGGCAGGGCCTACATCAATATCATAACGAAACGAGCAATGAAGATCGATGGCATTGAAAAGAACAAAGCCATAGCTGTTGATTCTGAAACCGGCGAGGTGTATTTCGGTACAACCGACGGACTATACAAATACGATCCAATAAATAACGAAGCTACAAATATCGGACTGTACAATGTGAATATAATGAAACTAGTCGTGAGGAACAACCAAATGTATTTGCTCGATGCCAACAATCATATGATTTACAAAGTCTACGACCACGGAAAGGCTACGATCAGAATGAGAAACGGGAAAACTGTGGTTGAATTCGACGTGGACGAGCGAGGCAATGTCCATTATGTCACGTTGTGCGGCTTATACTGCGCCATTGAGAACGGCGAAGTTATCAAGAACAAAGATTTGGATATCGTTAACAATTTTATTGTACACGAGGAGAAGACGTTTGCCGTCACCGAAGAAGGCTTGTATGATATCGATTGCGAGAATGGAACAGCAACGAGAGTTGCCCGTCTCGATTTCGCCCCTAGAAGCATGACTTTTGGTGATTATGGCGATATCTTTTATTCGCTCGACGACGCTATATACAGGTTAAGGCCAATAGTGTCTTATCAAGTTTACAATTTGTACGGGAGAAGTTGA
Protein
MLMKYLVMLIVASAEAKIRIYLTDTPLQIIGTRLYKKELLTDLYQAPKELTYDSSSRNLYFMYMDDSLQNSGRAYINIITKRAMKIDGIEKNKAIAVDSETGEVYFGTTDGLYKYDPINNEATNIGLYNVNIMKLVVRNNQMYLLDANNHMIYKVYDHGKATIRMRNGKTVVEFDVDERGNVHYVTLCGLYCAIENGEVIKNKDLDIVNNFIVHEEKTFAVTEEGLYDIDCENGTATRVARLDFAPRSMTFGDYGDIFYSLDDAIYRLRPIVSYQVYNLYGRS
Summary
Uniprot
H9JF83
A0A212EMS6
A0A2A4K8W2
A0A194QCP3
A0A2H1WB71
A0A0L7L3N6
+ More
A0A0N0PCV6 A0A2H1VWR3 A0A1E1WDT9 A9NPD0 A0A2A4JHI0 A0A2W1B6X3 A0A194Q748 A0A194Q2I5 A0A212EGN2 A0A0N1IBR9 A0A2H1X3L6 H9JF38 A0A3S2NVL2 A0A212FKW6 A0A0L7L740 A0A2W1BRP6 A0A2A4J3R5 A0A212F0W5 A0A2W1BAP6 A0A2A4J2B6 A0A2A4J225 A0A0L7LQ47 A0A0L7K2L6 A0A0N0PDM2 A0A194Q6N3 A0A2W1BT64 A0A2H1W986 A0A0L7LCK9 A0A212EMS2 A0A0L7L5A1 A0A212F574 A0A2A4JCK0 A0A194PKB7 S4PC69 A0A194R7A2 H9JCP0 A0A0L7K4L3 A0A0N1IJL9 H9JCN9 A0A0L7L7P2 H9JHT4 A0A2H1X0B0 I4DL32 A0A2A4J6D9
A0A0N0PCV6 A0A2H1VWR3 A0A1E1WDT9 A9NPD0 A0A2A4JHI0 A0A2W1B6X3 A0A194Q748 A0A194Q2I5 A0A212EGN2 A0A0N1IBR9 A0A2H1X3L6 H9JF38 A0A3S2NVL2 A0A212FKW6 A0A0L7L740 A0A2W1BRP6 A0A2A4J3R5 A0A212F0W5 A0A2W1BAP6 A0A2A4J2B6 A0A2A4J225 A0A0L7LQ47 A0A0L7K2L6 A0A0N0PDM2 A0A194Q6N3 A0A2W1BT64 A0A2H1W986 A0A0L7LCK9 A0A212EMS2 A0A0L7L5A1 A0A212F574 A0A2A4JCK0 A0A194PKB7 S4PC69 A0A194R7A2 H9JCP0 A0A0L7K4L3 A0A0N1IJL9 H9JCN9 A0A0L7L7P2 H9JHT4 A0A2H1X0B0 I4DL32 A0A2A4J6D9
EMBL
BABH01026891
AGBW02013802
OWR42794.1
NWSH01000023
PCG80650.1
PCG80651.1
+ More
KQ459386 KPJ01211.1 ODYU01007496 SOQ50313.1 JTDY01003236 KOB69904.1 KQ460423 KPJ14850.1 ODYU01004895 SOQ45238.1 GDQN01005929 JAT85125.1 EF083140 ABK22491.1 NWSH01001435 PCG71236.1 KZ150334 PZC71338.1 KPJ01209.1 KQ459562 KPI99756.1 AGBW02015065 OWR40643.1 KQ461158 KPJ08078.1 ODYU01013195 SOQ59889.1 BABH01026938 RSAL01000055 RVE49936.1 AGBW02007955 OWR54376.1 JTDY01002489 KOB71312.1 KZ149980 PZC75827.1 NWSH01003616 PCG66052.1 AGBW02011018 OWR47363.1 KZ150180 PZC72519.1 NWSH01004004 PCG65580.1 PCG66051.1 JTDY01000338 KOB77633.1 JTDY01014199 KOB52046.1 KQ460299 KPJ16155.1 KPJ01193.1 PZC75826.1 ODYU01007026 SOQ49402.1 JTDY01001660 KOB73227.1 OWR42795.1 JTDY01002898 KOB70521.1 AGBW02010227 OWR48887.1 NWSH01001998 PCG69476.1 KQ459602 KPI93453.1 GAIX01007995 JAA84565.1 KQ460615 KPJ13703.1 BABH01034622 JTDY01011636 KOB54879.1 KQ459895 KPJ19520.1 BABH01034621 KOB71311.1 BABH01029428 ODYU01012396 SOQ58682.1 AK402000 BAM18622.1 NWSH01003079 PCG66983.1
KQ459386 KPJ01211.1 ODYU01007496 SOQ50313.1 JTDY01003236 KOB69904.1 KQ460423 KPJ14850.1 ODYU01004895 SOQ45238.1 GDQN01005929 JAT85125.1 EF083140 ABK22491.1 NWSH01001435 PCG71236.1 KZ150334 PZC71338.1 KPJ01209.1 KQ459562 KPI99756.1 AGBW02015065 OWR40643.1 KQ461158 KPJ08078.1 ODYU01013195 SOQ59889.1 BABH01026938 RSAL01000055 RVE49936.1 AGBW02007955 OWR54376.1 JTDY01002489 KOB71312.1 KZ149980 PZC75827.1 NWSH01003616 PCG66052.1 AGBW02011018 OWR47363.1 KZ150180 PZC72519.1 NWSH01004004 PCG65580.1 PCG66051.1 JTDY01000338 KOB77633.1 JTDY01014199 KOB52046.1 KQ460299 KPJ16155.1 KPJ01193.1 PZC75826.1 ODYU01007026 SOQ49402.1 JTDY01001660 KOB73227.1 OWR42795.1 JTDY01002898 KOB70521.1 AGBW02010227 OWR48887.1 NWSH01001998 PCG69476.1 KQ459602 KPI93453.1 GAIX01007995 JAA84565.1 KQ460615 KPJ13703.1 BABH01034622 JTDY01011636 KOB54879.1 KQ459895 KPJ19520.1 BABH01034621 KOB71311.1 BABH01029428 ODYU01012396 SOQ58682.1 AK402000 BAM18622.1 NWSH01003079 PCG66983.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9JF83
A0A212EMS6
A0A2A4K8W2
A0A194QCP3
A0A2H1WB71
A0A0L7L3N6
+ More
A0A0N0PCV6 A0A2H1VWR3 A0A1E1WDT9 A9NPD0 A0A2A4JHI0 A0A2W1B6X3 A0A194Q748 A0A194Q2I5 A0A212EGN2 A0A0N1IBR9 A0A2H1X3L6 H9JF38 A0A3S2NVL2 A0A212FKW6 A0A0L7L740 A0A2W1BRP6 A0A2A4J3R5 A0A212F0W5 A0A2W1BAP6 A0A2A4J2B6 A0A2A4J225 A0A0L7LQ47 A0A0L7K2L6 A0A0N0PDM2 A0A194Q6N3 A0A2W1BT64 A0A2H1W986 A0A0L7LCK9 A0A212EMS2 A0A0L7L5A1 A0A212F574 A0A2A4JCK0 A0A194PKB7 S4PC69 A0A194R7A2 H9JCP0 A0A0L7K4L3 A0A0N1IJL9 H9JCN9 A0A0L7L7P2 H9JHT4 A0A2H1X0B0 I4DL32 A0A2A4J6D9
A0A0N0PCV6 A0A2H1VWR3 A0A1E1WDT9 A9NPD0 A0A2A4JHI0 A0A2W1B6X3 A0A194Q748 A0A194Q2I5 A0A212EGN2 A0A0N1IBR9 A0A2H1X3L6 H9JF38 A0A3S2NVL2 A0A212FKW6 A0A0L7L740 A0A2W1BRP6 A0A2A4J3R5 A0A212F0W5 A0A2W1BAP6 A0A2A4J2B6 A0A2A4J225 A0A0L7LQ47 A0A0L7K2L6 A0A0N0PDM2 A0A194Q6N3 A0A2W1BT64 A0A2H1W986 A0A0L7LCK9 A0A212EMS2 A0A0L7L5A1 A0A212F574 A0A2A4JCK0 A0A194PKB7 S4PC69 A0A194R7A2 H9JCP0 A0A0L7K4L3 A0A0N1IJL9 H9JCN9 A0A0L7L7P2 H9JHT4 A0A2H1X0B0 I4DL32 A0A2A4J6D9
PDB
5FWW
E-value=0.000177583,
Score=105
Ontologies
PANTHER
Topology
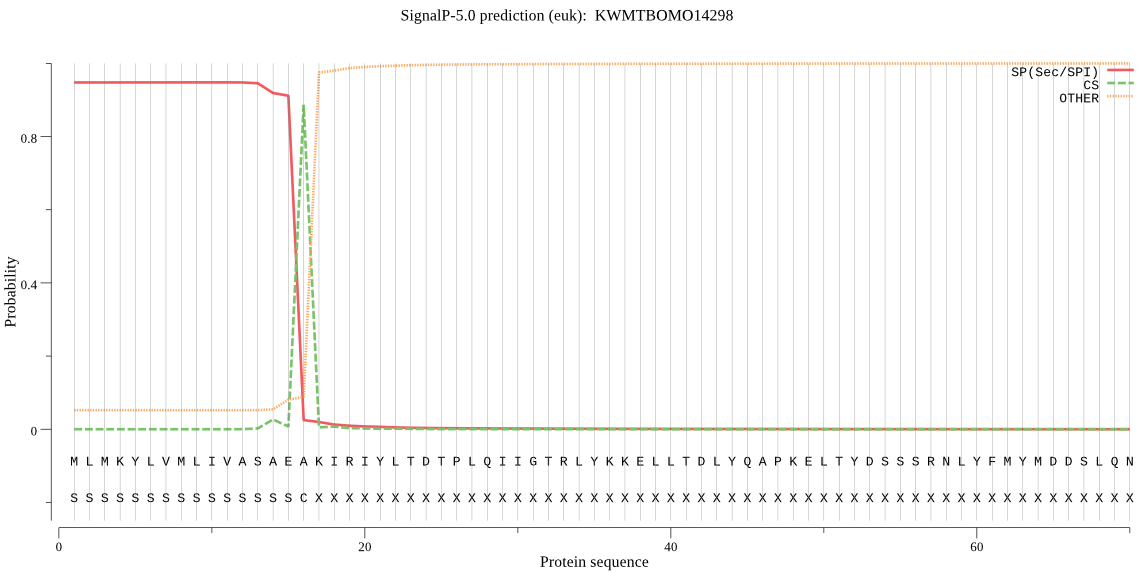
SignalP
Position: 1 - 16,
Likelihood: 0.946815
Length:
283
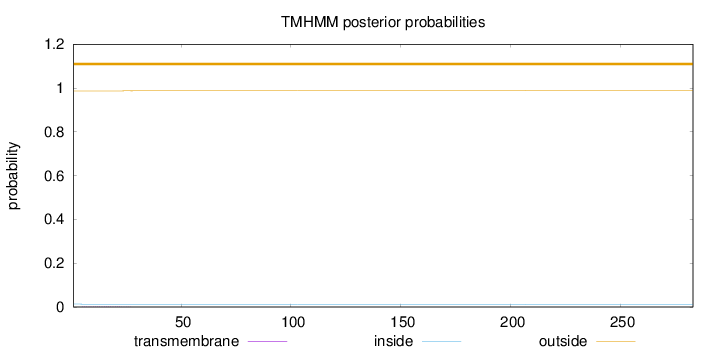
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03017
Exp number, first 60 AAs:
0.02775
Total prob of N-in:
0.01371
outside
1 - 283
Population Genetic Test Statistics
Pi
300.674859
Theta
215.970898
Tajima's D
1.093245
CLR
0.158245
CSRT
0.685165741712914
Interpretation
Uncertain
