Gene
KWMTBOMO14297 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008179
Annotation
Ommochrome-binding_protein_[Operophtera_brumata]
Full name
Ommochrome-binding protein
Alternative Name
YCP
Location in the cell
Extracellular Reliability : 1.378 PlasmaMembrane Reliability : 1.261
Sequence
CDS
ATGAAGTTGTTCTTGTTAATGACAAGCGTAATCGTGACTCAAGCGGGAACCCTCAGAGGGGAGTGCAAAGGCGTCGAAATCAACGGGGCGACGTACGAAGAGGAGATGCTCTACGAAAATCTGGACCGCCCTTACCTTCTGACCGTGGACTACAGCACCAACGACCTGTACTTCAGCTATAGCATAGACAACGACGAGGACAGCTCCGCAACGGCAAGATATAATTTGGACACGAAGAAGTTTACCACCATCGAGGGCGTGAATAATGGCTTCGCGCAGACTGTGGACCAAACCACGCAGACCGTGTACATAGGCGGAAGCGACGGTCTATATCGATACGACAAGGATACCAAAAGAGCGGTATTAATAGGAGCACCTAGTATTAACATATGGACCATATTCTACAAAGACGTGTTGTATTTCTCAACGTACCCCTCACACATTCTCTACACGTTTTCAAACGGTCAGATCACCAGGTTCCCAGATCTAGAAGACACTAGAGTAGACAACTTTATTATAGACAATGATGATTACATTTTCTTCACGAATACCTCTGGTCTTTACAGACAGAAAAAGGGTACTAAAGACGCGGTTTTGTACGAGGTGTTTGAAGATTTCGGTGTGAGAGGATTTGCCTTAGATGCACAGGGCGTGGTGCACGCTTGCTTCCAAAACGGCGTATACTCTCTGAATAACACCGAGGTCGGCCTAAAGAAGATTGTAAGCGTGGATGATGCGTTCGGCGTCGCTTTTGATAAAGACAATAATCTTATTTATTCTGATGCTAGAAGAGTCATTCGACTGAAACCTACAAAGGATTGTTAA
Protein
MKLFLLMTSVIVTQAGTLRGECKGVEINGATYEEEMLYENLDRPYLLTVDYSTNDLYFSYSIDNDEDSSATARYNLDTKKFTTIEGVNNGFAQTVDQTTQTVYIGGSDGLYRYDKDTKRAVLIGAPSINIWTIFYKDVLYFSTYPSHILYTFSNGQITRFPDLEDTRVDNFIIDNDDYIFFTNTSGLYRQKKGTKDAVLYEVFEDFGVRGFALDAQGVVHACFQNGVYSLNNTEVGLKKIVSVDDAFGVAFDKDNNLIYSDARRVIRLKPTKDC
Summary
Subunit
Monomer.
Keywords
Direct protein sequencing
Glycoprotein
Signal
Feature
chain Ommochrome-binding protein
Uniprot
H9JF82
A0A0L7L3N6
A0A0N0PCV6
A0A194Q748
A0A212EMS2
A0A0N1PJI9
+ More
A0A212EGN2 A0A194Q763 A9NPD0 A0A1E1WDT9 I4DL32 A0A0L7LCK9 A0A0L7K2L6 S4PC69 H9BE66 A0A0L7LDZ5 P31420 A0A0L7L740 A0A2W1BID2 A0A2A4JCK0 A0A0L7K4L3 A0A212FC69 A0A194R7A2 A0A2H1VWR3 A0A212F0W5 A0A212FKW6 A0A194PKB7 A0A0L7L7P2 A0A0L7L5A1 A0A2W1BW95 A0A2A4JHI0 A0A0N1IBR9 Q9NDA4 Q5PY94 A0A0L7L3P1 H9JMK0 A0A194Q2I5 A0A2A4K3H5 H9JCP0 A0A0N0PE93 Q5PY95 A0A2W1BAP6 H9JDD0 Q5PY92 Q5PY91 Q5PY93 A0A2H1X0B0 A0A2A4K6L9 H9JHT4 A0A0N1IJL9 H9JCN9 A0A212EJ06 A0A212EMV3 A0A2A4J2B6 A0A2W1B6X3 A0A0N1PGN6 H9JF79 A0A0L7L941 A0A194Q899 A0A2W1BWL3 A0A2H1X3L6 A0A0L7L354 A0A3S2KYT5 A0A3S2PB03 A0A194QD66 A0A194REA4 A0A1E1WC11 A0A2H1V2A2 A0A0L7KVE0
A0A212EGN2 A0A194Q763 A9NPD0 A0A1E1WDT9 I4DL32 A0A0L7LCK9 A0A0L7K2L6 S4PC69 H9BE66 A0A0L7LDZ5 P31420 A0A0L7L740 A0A2W1BID2 A0A2A4JCK0 A0A0L7K4L3 A0A212FC69 A0A194R7A2 A0A2H1VWR3 A0A212F0W5 A0A212FKW6 A0A194PKB7 A0A0L7L7P2 A0A0L7L5A1 A0A2W1BW95 A0A2A4JHI0 A0A0N1IBR9 Q9NDA4 Q5PY94 A0A0L7L3P1 H9JMK0 A0A194Q2I5 A0A2A4K3H5 H9JCP0 A0A0N0PE93 Q5PY95 A0A2W1BAP6 H9JDD0 Q5PY92 Q5PY91 Q5PY93 A0A2H1X0B0 A0A2A4K6L9 H9JHT4 A0A0N1IJL9 H9JCN9 A0A212EJ06 A0A212EMV3 A0A2A4J2B6 A0A2W1B6X3 A0A0N1PGN6 H9JF79 A0A0L7L941 A0A194Q899 A0A2W1BWL3 A0A2H1X3L6 A0A0L7L354 A0A3S2KYT5 A0A3S2PB03 A0A194QD66 A0A194REA4 A0A1E1WC11 A0A2H1V2A2 A0A0L7KVE0
Pubmed
EMBL
BABH01026891
JTDY01003236
KOB69904.1
KQ460423
KPJ14850.1
KQ459386
+ More
KPJ01209.1 AGBW02013802 OWR42795.1 KPJ14832.1 AGBW02015065 OWR40643.1 KPJ01224.1 EF083140 ABK22491.1 GDQN01005929 JAT85125.1 AK402000 BAM18622.1 JTDY01001660 KOB73227.1 JTDY01014199 KOB52046.1 GAIX01007995 JAA84565.1 JQ003184 AFC35301.1 JTDY01001487 KOB73748.1 L00975 JTDY01002489 KOB71312.1 KZ150180 PZC72520.1 NWSH01001998 PCG69476.1 JTDY01011636 KOB54879.1 AGBW02009212 OWR51334.1 KQ460615 KPJ13703.1 ODYU01004895 SOQ45238.1 AGBW02011018 OWR47363.1 AGBW02007955 OWR54376.1 KQ459602 KPI93453.1 KOB71311.1 JTDY01002898 KOB70521.1 KZ149928 PZC77477.1 NWSH01001435 PCG71236.1 KQ461158 KPJ08078.1 AF169311 AAF89672.1 AY819652 AAV68385.1 JTDY01003234 KOB69909.1 BABH01016630 KQ459562 KPI99756.1 NWSH01000191 PCG78576.1 BABH01034622 KQ459895 KPJ19519.1 AY819651 AAV68384.1 PZC72519.1 BABH01030111 AY819654 AAV68387.1 AY819655 AAV68388.1 AY819653 AAV68386.1 ODYU01012396 SOQ58682.1 NWSH01000114 PCG79413.1 BABH01029428 KPJ19520.1 BABH01034621 AGBW02014547 OWR41451.1 OWR42797.1 NWSH01004004 PCG65580.1 KZ150334 PZC71338.1 KPJ14845.1 BABH01026888 JTDY01002143 KOB72012.1 KPJ01215.1 KZ149968 PZC76133.1 ODYU01013195 SOQ59889.1 KOB69902.1 RSAL01000448 RVE41684.1 RSAL01000133 RVE46334.1 KQ459337 KPJ01396.1 KQ460297 KPJ16163.1 GDQN01006536 JAT84518.1 ODYU01000350 SOQ34961.1 JTDY01005429 KOB67004.1
KPJ01209.1 AGBW02013802 OWR42795.1 KPJ14832.1 AGBW02015065 OWR40643.1 KPJ01224.1 EF083140 ABK22491.1 GDQN01005929 JAT85125.1 AK402000 BAM18622.1 JTDY01001660 KOB73227.1 JTDY01014199 KOB52046.1 GAIX01007995 JAA84565.1 JQ003184 AFC35301.1 JTDY01001487 KOB73748.1 L00975 JTDY01002489 KOB71312.1 KZ150180 PZC72520.1 NWSH01001998 PCG69476.1 JTDY01011636 KOB54879.1 AGBW02009212 OWR51334.1 KQ460615 KPJ13703.1 ODYU01004895 SOQ45238.1 AGBW02011018 OWR47363.1 AGBW02007955 OWR54376.1 KQ459602 KPI93453.1 KOB71311.1 JTDY01002898 KOB70521.1 KZ149928 PZC77477.1 NWSH01001435 PCG71236.1 KQ461158 KPJ08078.1 AF169311 AAF89672.1 AY819652 AAV68385.1 JTDY01003234 KOB69909.1 BABH01016630 KQ459562 KPI99756.1 NWSH01000191 PCG78576.1 BABH01034622 KQ459895 KPJ19519.1 AY819651 AAV68384.1 PZC72519.1 BABH01030111 AY819654 AAV68387.1 AY819655 AAV68388.1 AY819653 AAV68386.1 ODYU01012396 SOQ58682.1 NWSH01000114 PCG79413.1 BABH01029428 KPJ19520.1 BABH01034621 AGBW02014547 OWR41451.1 OWR42797.1 NWSH01004004 PCG65580.1 KZ150334 PZC71338.1 KPJ14845.1 BABH01026888 JTDY01002143 KOB72012.1 KPJ01215.1 KZ149968 PZC76133.1 ODYU01013195 SOQ59889.1 KOB69902.1 RSAL01000448 RVE41684.1 RSAL01000133 RVE46334.1 KQ459337 KPJ01396.1 KQ460297 KPJ16163.1 GDQN01006536 JAT84518.1 ODYU01000350 SOQ34961.1 JTDY01005429 KOB67004.1
Proteomes
Interpro
IPR015943
WD40/YVTN_repeat-like_dom_sf
+ More
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR017850 Alkaline_phosphatase_core_sf
IPR004245 DUF229
IPR011047 Quinoprotein_ADH-like_supfam
IPR001958 Tet-R_TetA/multi-R_MdtG
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR011042 6-blade_b-propeller_TolB-like
IPR036574 Scorpion_toxin-like_sf
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR017850 Alkaline_phosphatase_core_sf
IPR004245 DUF229
IPR011047 Quinoprotein_ADH-like_supfam
IPR001958 Tet-R_TetA/multi-R_MdtG
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR011042 6-blade_b-propeller_TolB-like
IPR036574 Scorpion_toxin-like_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JF82
A0A0L7L3N6
A0A0N0PCV6
A0A194Q748
A0A212EMS2
A0A0N1PJI9
+ More
A0A212EGN2 A0A194Q763 A9NPD0 A0A1E1WDT9 I4DL32 A0A0L7LCK9 A0A0L7K2L6 S4PC69 H9BE66 A0A0L7LDZ5 P31420 A0A0L7L740 A0A2W1BID2 A0A2A4JCK0 A0A0L7K4L3 A0A212FC69 A0A194R7A2 A0A2H1VWR3 A0A212F0W5 A0A212FKW6 A0A194PKB7 A0A0L7L7P2 A0A0L7L5A1 A0A2W1BW95 A0A2A4JHI0 A0A0N1IBR9 Q9NDA4 Q5PY94 A0A0L7L3P1 H9JMK0 A0A194Q2I5 A0A2A4K3H5 H9JCP0 A0A0N0PE93 Q5PY95 A0A2W1BAP6 H9JDD0 Q5PY92 Q5PY91 Q5PY93 A0A2H1X0B0 A0A2A4K6L9 H9JHT4 A0A0N1IJL9 H9JCN9 A0A212EJ06 A0A212EMV3 A0A2A4J2B6 A0A2W1B6X3 A0A0N1PGN6 H9JF79 A0A0L7L941 A0A194Q899 A0A2W1BWL3 A0A2H1X3L6 A0A0L7L354 A0A3S2KYT5 A0A3S2PB03 A0A194QD66 A0A194REA4 A0A1E1WC11 A0A2H1V2A2 A0A0L7KVE0
A0A212EGN2 A0A194Q763 A9NPD0 A0A1E1WDT9 I4DL32 A0A0L7LCK9 A0A0L7K2L6 S4PC69 H9BE66 A0A0L7LDZ5 P31420 A0A0L7L740 A0A2W1BID2 A0A2A4JCK0 A0A0L7K4L3 A0A212FC69 A0A194R7A2 A0A2H1VWR3 A0A212F0W5 A0A212FKW6 A0A194PKB7 A0A0L7L7P2 A0A0L7L5A1 A0A2W1BW95 A0A2A4JHI0 A0A0N1IBR9 Q9NDA4 Q5PY94 A0A0L7L3P1 H9JMK0 A0A194Q2I5 A0A2A4K3H5 H9JCP0 A0A0N0PE93 Q5PY95 A0A2W1BAP6 H9JDD0 Q5PY92 Q5PY91 Q5PY93 A0A2H1X0B0 A0A2A4K6L9 H9JHT4 A0A0N1IJL9 H9JCN9 A0A212EJ06 A0A212EMV3 A0A2A4J2B6 A0A2W1B6X3 A0A0N1PGN6 H9JF79 A0A0L7L941 A0A194Q899 A0A2W1BWL3 A0A2H1X3L6 A0A0L7L354 A0A3S2KYT5 A0A3S2PB03 A0A194QD66 A0A194REA4 A0A1E1WC11 A0A2H1V2A2 A0A0L7KVE0
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.985911
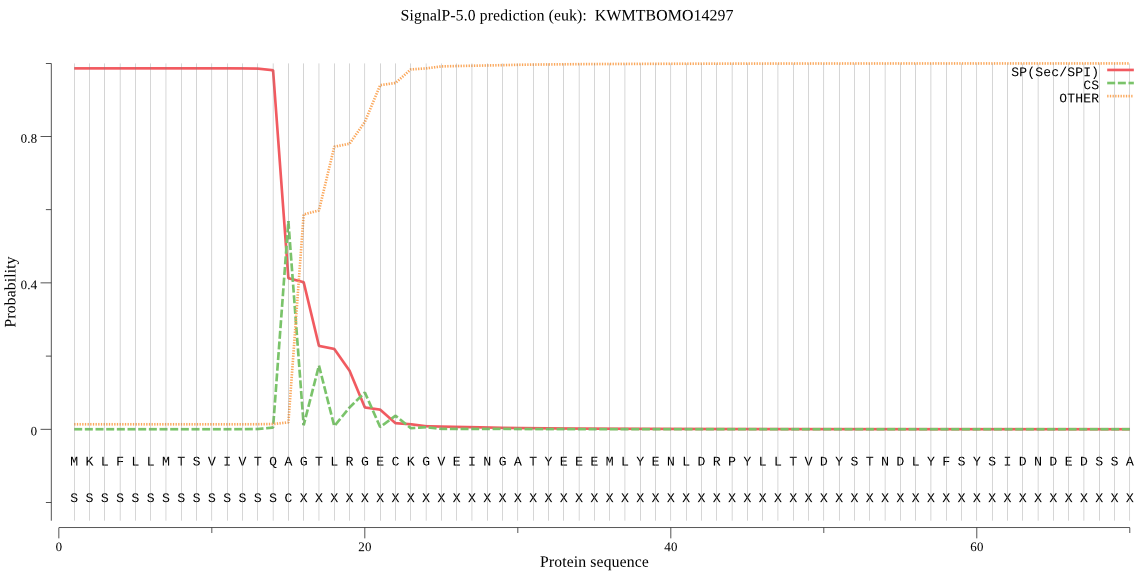
Length:
274
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.597159999999998
Exp number, first 60 AAs:
0.00174
Total prob of N-in:
0.03180
outside
1 - 274

Population Genetic Test Statistics
Pi
209.214448
Theta
182.194213
Tajima's D
0.463317
CLR
0.99699
CSRT
0.505974701264937
Interpretation
Uncertain
