Gene
KWMTBOMO14293
Pre Gene Modal
BGIBMGA008144
Annotation
PREDICTED:_uncharacterized_protein_LOC101744639_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.054 Mitochondrial Reliability : 1.578
Sequence
CDS
ATGCACCGGACGTCTTATCCTTTAGGCCACGACGACTTTCAAAAGGACATAACCTGTACGTACAGAGACATCATATACCACAGTGACAACGAGTACGTATTGGGACCGCCGACTGACGTCCGGGATGGCGGCGTGTACCACCTGACGAGAAGCGACCACGTCAAGATCAAATGCGTAGGCACGCATCTGCATAGCATCCTTCCGTCGAGATGGTCAGGTCATTCGCTGGGATTCCGGTCGACGGTGGCGGCGCCGCCCCCCGACCGCGATGACGCTCTCGACGTGCTCATATTCGGGTTCGATTCCACGTCGAAGAACGGCTTCATAAGGAGTATGCCGAAGAGCTACAAATACCTGACCGAGGAGCTGAATGGCTACGTTCTAGACGGGTACGGCATACTCGGTGACGGCACGCCTGCCGCCTTATTCCCCATACTCACGGGCAAGAACGAGTTGGAGCTACCAGACGTCAGGAAGAAGACGAAGAACGGCAACACCTTAGACTGTATGCCGTTCATATTCAACCAACTAAAGGGGGACGGTTACCGCACGGCATATTTCGAGGACATGCCCTGGATAGGAACGTTTCAGTACCGGTTTAATGGTTTCCTCCATCAGCCGGCCGATCATTACTTGAGGGCATTCTACCTCGAGGAGTCGAACAGAGGCAAGAAGTGGTGGAGGGGCAGCAACAGCAAGTACTGCATAGGAGACACTCCTCAGTACAAGCTAATGATGAACATTACCGATCAGTTCTTTCGTTTGGACGGCAAGAAGTTTTGCTTCACCTTCATAGCCGACATAACTCATGATGACATCAACATGATATCGACAGCTGACGATGACACTGTCGACTTCTTGAAGCTGTTCAAGGAACAAGGAAGGCTTGAGAACACCATGCTATTCGTGATGGGCGACCACGGACCCAGGTACTCGAAGGTCCGCAACACGCTGCAAGGCAAGCTGGAGGAGCGTCTACCGCTGATGGTGGTGGTCCCGGCGGCGCGGCTGGTGCGCGCGCGGCCCGCCGTGCTCGAAGCGCTGCGACACAACCGCGCCGCGCTCACCACGCCGCACGACGTGCACGCCACGCTGCTCGACGCGCTGCGCCGCCCCGCCCCGCCCCCCGCCTACACCGTGCCCGGCGCCGACCTGCCGCGCGCGCTCTCCCTGCTGCAGCCGATCCCGCGGAACCGGTCGTGCAGCGAGGCGGGCATTGAGCCGCACTGGTGCGCGTGCATGCGCTGGAGCCCCGTGCCCCCCGCCGACCCGCTGCACGCCGCCGCCCCCCGCGCGCTACTGCAATACATCAACACGCTCACCGAGCCCAAAAGAGCGTCGTGCGTCCCCCGCACGCTGTCCGCGGTGCACTGGGTGCTGAGGGCCCACGCCAACAGTAGGGTGCTCTCCTTCATCGAGGCCAAGGACGCGGACGGCTACGTGGGGAAGTTCGGGGCCAAGCTCAAGGTCAACAGGGAGAACTTGCAGGTCAAGATCTCCGTCGGTCCCGGCGAGGGGATATACGAGGCCTCGCTCGCGTACCTGAGAAAGGAGGACAAGTTCCTCATCGAGTCCAGAGACATATCCAGGACGAACGCGTACAACGACGAGCCCAGCTGCATCAGCGCCACGCACCCCCACCTCAACAAGTACTGTTACTGCAAAATATAG
Protein
MHRTSYPLGHDDFQKDITCTYRDIIYHSDNEYVLGPPTDVRDGGVYHLTRSDHVKIKCVGTHLHSILPSRWSGHSLGFRSTVAAPPPDRDDALDVLIFGFDSTSKNGFIRSMPKSYKYLTEELNGYVLDGYGILGDGTPAALFPILTGKNELELPDVRKKTKNGNTLDCMPFIFNQLKGDGYRTAYFEDMPWIGTFQYRFNGFLHQPADHYLRAFYLEESNRGKKWWRGSNSKYCIGDTPQYKLMMNITDQFFRLDGKKFCFTFIADITHDDINMISTADDDTVDFLKLFKEQGRLENTMLFVMGDHGPRYSKVRNTLQGKLEERLPLMVVVPAARLVRARPAVLEALRHNRAALTTPHDVHATLLDALRRPAPPPAYTVPGADLPRALSLLQPIPRNRSCSEAGIEPHWCACMRWSPVPPADPLHAAAPRALLQYINTLTEPKRASCVPRTLSAVHWVLRAHANSRVLSFIEAKDADGYVGKFGAKLKVNRENLQVKISVGPGEGIYEASLAYLRKEDKFLIESRDISRTNAYNDEPSCISATHPHLNKYCYCKI
Summary
Uniprot
A0A194Q753
A0A212ERQ6
A0A0N1IA58
A0A0N1IEY9
A0A0L7LA27
A0A212EMV1
+ More
A0A2H1VJH0 A0A2A4K8N2 D6WJE5 A0A0T6AXB0 A0A336LZA2 A0A194Q763 A0A2R7WC52 B4LZX9 A0A0M9A6K9 A0A182VQ70 A0A0L7QSC4 A0A1Y1MZ42 A0A182RPN3 A0A182QRZ4 A0A087ZVH2 A0A0A9XK59 A0A182FNS2 W5JNU8 A0A182N4Q8 Q7QAP4 A0A154P2D5 A0A182HZC6 A0A182WVR2 A0A182KXI8 A0A182VB63 A0A182P929 A0A310SK20 A0A182IU05 A0A182TG03 A0A182Y7C0 A0A182ME46 A0A084VHA2 B4K5U0 B4GLC4 Q294A6 W8AR20 B0XIU0 A0A1S4G777 A0A1Q3F0A8 B4JGD9 A0A0A9XQ18 A0A1B6K7B3 A0A2H1VSZ8 Q0IEE1 A0A1B0ET90 Q95TI3 Q9VF91 A0A3B0KEN1 A0A067QPR6 E0VYN6 A0A1B0DN89 A0A1B6DNU0 A0A182JRJ7 T1IUT3 T1IDN1 A0A3S3PZD6 A0A1I8Q2Q8 K1RB06 A0A1J1INU8 A0A2J7R5X9 E2B718 K1Q1E6 A0A2J7R5X6 U4U1C6 N6U6B5 U4UTH7 K7IY54 A0A1I8MTR3 A0A2W1BM30 A0A151IPS5 A0A026X1E7 A0A087T143 A0A2P8YAH3 A0A195DZ05 A0A151X0G3 A0A2L2YJ02 A0A195EWS7 F4W5Q6 A0A195BTB1 A0A1B0AJA1 A0A1A9V2J2 E1ZY92 A0A1B0GDU8 A0A2A3ESR3 X1WJK4 A0A1A9X438 A0A3S3NU32 A0A0L8HAX3 A0A1I8Q2R0 V4BRA1 A0A1I8Q2N3
A0A2H1VJH0 A0A2A4K8N2 D6WJE5 A0A0T6AXB0 A0A336LZA2 A0A194Q763 A0A2R7WC52 B4LZX9 A0A0M9A6K9 A0A182VQ70 A0A0L7QSC4 A0A1Y1MZ42 A0A182RPN3 A0A182QRZ4 A0A087ZVH2 A0A0A9XK59 A0A182FNS2 W5JNU8 A0A182N4Q8 Q7QAP4 A0A154P2D5 A0A182HZC6 A0A182WVR2 A0A182KXI8 A0A182VB63 A0A182P929 A0A310SK20 A0A182IU05 A0A182TG03 A0A182Y7C0 A0A182ME46 A0A084VHA2 B4K5U0 B4GLC4 Q294A6 W8AR20 B0XIU0 A0A1S4G777 A0A1Q3F0A8 B4JGD9 A0A0A9XQ18 A0A1B6K7B3 A0A2H1VSZ8 Q0IEE1 A0A1B0ET90 Q95TI3 Q9VF91 A0A3B0KEN1 A0A067QPR6 E0VYN6 A0A1B0DN89 A0A1B6DNU0 A0A182JRJ7 T1IUT3 T1IDN1 A0A3S3PZD6 A0A1I8Q2Q8 K1RB06 A0A1J1INU8 A0A2J7R5X9 E2B718 K1Q1E6 A0A2J7R5X6 U4U1C6 N6U6B5 U4UTH7 K7IY54 A0A1I8MTR3 A0A2W1BM30 A0A151IPS5 A0A026X1E7 A0A087T143 A0A2P8YAH3 A0A195DZ05 A0A151X0G3 A0A2L2YJ02 A0A195EWS7 F4W5Q6 A0A195BTB1 A0A1B0AJA1 A0A1A9V2J2 E1ZY92 A0A1B0GDU8 A0A2A3ESR3 X1WJK4 A0A1A9X438 A0A3S3NU32 A0A0L8HAX3 A0A1I8Q2R0 V4BRA1 A0A1I8Q2N3
Pubmed
26354079
22118469
26227816
18362917
19820115
17994087
+ More
28004739 25401762 26823975 20920257 23761445 12364791 20966253 25244985 24438588 15632085 24495485 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 20566863 22992520 20798317 23537049 20075255 25315136 28756777 24508170 30249741 29403074 26561354 21719571 23254933
28004739 25401762 26823975 20920257 23761445 12364791 20966253 25244985 24438588 15632085 24495485 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 20566863 22992520 20798317 23537049 20075255 25315136 28756777 24508170 30249741 29403074 26561354 21719571 23254933
EMBL
KQ459386
KPJ01214.1
AGBW02012951
OWR44178.1
KQ460423
KPJ14844.1
+ More
KPJ14833.1 JTDY01002045 KOB72244.1 AGBW02013802 OWR42796.1 ODYU01002890 SOQ40941.1 NWSH01000023 PCG80645.1 KQ971342 EFA03884.1 LJIG01022597 KRT79734.1 UFQS01000197 UFQT01000197 SSX01187.1 SSX21567.1 KPJ01224.1 KK854603 PTY17274.1 CH940650 EDW67207.1 KQ435736 KOX76983.1 KQ414768 KOC61371.1 GEZM01017097 GEZM01017096 GEZM01017095 GEZM01017094 JAV90932.1 AXCN02000015 GBHO01024436 GBHO01024431 GDHC01016508 GDHC01005499 JAG19168.1 JAG19173.1 JAQ02121.1 JAQ13130.1 ADMH02001019 ETN64424.1 AAAB01008888 EAA08955.5 KQ434804 KZC06099.1 APCN01005540 KQ762207 OAD56073.1 AXCM01000623 ATLV01013150 KE524842 KFB37346.1 CH933806 EDW15152.1 CH479185 EDW38348.1 CM000070 EAL29058.1 GAMC01019582 JAB86973.1 DS233377 EDS29681.1 GFDL01014055 JAV20990.1 CH916369 EDV92608.1 GBHO01024434 GBHO01024433 JAG19170.1 JAG19171.1 GECU01000368 JAT07339.1 ODYU01004246 SOQ43918.1 CH477695 EAT37158.1 AJWK01005593 AJWK01005594 AY058755 AAL13984.1 AE014297 BT150241 AAF55169.1 AGV77143.1 OUUW01000007 SPP83511.1 KK853083 KDR11645.1 DS235845 EEB18492.1 AJVK01017387 GEDC01009960 JAS27338.1 JH431556 ACPB03002176 NCKU01010949 RWS00604.1 JH815790 EKC38410.1 CVRI01000057 CRL01832.1 NEVH01006987 PNF36237.1 GL446098 EFN88497.1 JH816402 EKC22605.1 PNF36236.1 KB630727 ERL83825.1 APGK01040621 KB740984 ENN76196.1 KB632357 ERL93435.1 KZ149968 PZC76132.1 KQ976812 KYN08157.1 KK107039 QOIP01000006 EZA61841.1 RLU22184.1 KK112918 KFM58832.1 PYGN01000755 PSN41256.1 KQ980050 KYN18061.1 KQ982630 KYQ53420.1 IAAA01022626 IAAA01022627 LAA07235.1 KQ981953 KYN32339.1 GL887680 EGI70467.1 KQ976417 KYM89671.1 GL435204 EFN73791.1 CCAG010012028 KZ288193 PBC34169.1 ABLF02024211 ABLF02024212 ABLF02024216 ABLF02024219 NCKU01008508 RWS01853.1 KQ418645 KOF86431.1 KB202283 ESO91374.1
KPJ14833.1 JTDY01002045 KOB72244.1 AGBW02013802 OWR42796.1 ODYU01002890 SOQ40941.1 NWSH01000023 PCG80645.1 KQ971342 EFA03884.1 LJIG01022597 KRT79734.1 UFQS01000197 UFQT01000197 SSX01187.1 SSX21567.1 KPJ01224.1 KK854603 PTY17274.1 CH940650 EDW67207.1 KQ435736 KOX76983.1 KQ414768 KOC61371.1 GEZM01017097 GEZM01017096 GEZM01017095 GEZM01017094 JAV90932.1 AXCN02000015 GBHO01024436 GBHO01024431 GDHC01016508 GDHC01005499 JAG19168.1 JAG19173.1 JAQ02121.1 JAQ13130.1 ADMH02001019 ETN64424.1 AAAB01008888 EAA08955.5 KQ434804 KZC06099.1 APCN01005540 KQ762207 OAD56073.1 AXCM01000623 ATLV01013150 KE524842 KFB37346.1 CH933806 EDW15152.1 CH479185 EDW38348.1 CM000070 EAL29058.1 GAMC01019582 JAB86973.1 DS233377 EDS29681.1 GFDL01014055 JAV20990.1 CH916369 EDV92608.1 GBHO01024434 GBHO01024433 JAG19170.1 JAG19171.1 GECU01000368 JAT07339.1 ODYU01004246 SOQ43918.1 CH477695 EAT37158.1 AJWK01005593 AJWK01005594 AY058755 AAL13984.1 AE014297 BT150241 AAF55169.1 AGV77143.1 OUUW01000007 SPP83511.1 KK853083 KDR11645.1 DS235845 EEB18492.1 AJVK01017387 GEDC01009960 JAS27338.1 JH431556 ACPB03002176 NCKU01010949 RWS00604.1 JH815790 EKC38410.1 CVRI01000057 CRL01832.1 NEVH01006987 PNF36237.1 GL446098 EFN88497.1 JH816402 EKC22605.1 PNF36236.1 KB630727 ERL83825.1 APGK01040621 KB740984 ENN76196.1 KB632357 ERL93435.1 KZ149968 PZC76132.1 KQ976812 KYN08157.1 KK107039 QOIP01000006 EZA61841.1 RLU22184.1 KK112918 KFM58832.1 PYGN01000755 PSN41256.1 KQ980050 KYN18061.1 KQ982630 KYQ53420.1 IAAA01022626 IAAA01022627 LAA07235.1 KQ981953 KYN32339.1 GL887680 EGI70467.1 KQ976417 KYM89671.1 GL435204 EFN73791.1 CCAG010012028 KZ288193 PBC34169.1 ABLF02024211 ABLF02024212 ABLF02024216 ABLF02024219 NCKU01008508 RWS01853.1 KQ418645 KOF86431.1 KB202283 ESO91374.1
Proteomes
UP000053268
UP000007151
UP000053240
UP000037510
UP000218220
UP000007266
+ More
UP000008792 UP000053105 UP000075920 UP000053825 UP000075900 UP000075886 UP000005203 UP000069272 UP000000673 UP000075884 UP000007062 UP000076502 UP000075840 UP000076407 UP000075882 UP000075903 UP000075885 UP000075880 UP000075902 UP000076408 UP000075883 UP000030765 UP000009192 UP000008744 UP000001819 UP000002320 UP000001070 UP000008820 UP000092461 UP000000803 UP000268350 UP000027135 UP000009046 UP000092462 UP000075881 UP000015103 UP000285301 UP000095300 UP000005408 UP000183832 UP000235965 UP000008237 UP000030742 UP000019118 UP000002358 UP000095301 UP000078542 UP000053097 UP000279307 UP000054359 UP000245037 UP000078492 UP000075809 UP000078541 UP000007755 UP000078540 UP000092445 UP000078200 UP000000311 UP000092444 UP000242457 UP000007819 UP000091820 UP000053454 UP000030746
UP000008792 UP000053105 UP000075920 UP000053825 UP000075900 UP000075886 UP000005203 UP000069272 UP000000673 UP000075884 UP000007062 UP000076502 UP000075840 UP000076407 UP000075882 UP000075903 UP000075885 UP000075880 UP000075902 UP000076408 UP000075883 UP000030765 UP000009192 UP000008744 UP000001819 UP000002320 UP000001070 UP000008820 UP000092461 UP000000803 UP000268350 UP000027135 UP000009046 UP000092462 UP000075881 UP000015103 UP000285301 UP000095300 UP000005408 UP000183832 UP000235965 UP000008237 UP000030742 UP000019118 UP000002358 UP000095301 UP000078542 UP000053097 UP000279307 UP000054359 UP000245037 UP000078492 UP000075809 UP000078541 UP000007755 UP000078540 UP000092445 UP000078200 UP000000311 UP000092444 UP000242457 UP000007819 UP000091820 UP000053454 UP000030746
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194Q753
A0A212ERQ6
A0A0N1IA58
A0A0N1IEY9
A0A0L7LA27
A0A212EMV1
+ More
A0A2H1VJH0 A0A2A4K8N2 D6WJE5 A0A0T6AXB0 A0A336LZA2 A0A194Q763 A0A2R7WC52 B4LZX9 A0A0M9A6K9 A0A182VQ70 A0A0L7QSC4 A0A1Y1MZ42 A0A182RPN3 A0A182QRZ4 A0A087ZVH2 A0A0A9XK59 A0A182FNS2 W5JNU8 A0A182N4Q8 Q7QAP4 A0A154P2D5 A0A182HZC6 A0A182WVR2 A0A182KXI8 A0A182VB63 A0A182P929 A0A310SK20 A0A182IU05 A0A182TG03 A0A182Y7C0 A0A182ME46 A0A084VHA2 B4K5U0 B4GLC4 Q294A6 W8AR20 B0XIU0 A0A1S4G777 A0A1Q3F0A8 B4JGD9 A0A0A9XQ18 A0A1B6K7B3 A0A2H1VSZ8 Q0IEE1 A0A1B0ET90 Q95TI3 Q9VF91 A0A3B0KEN1 A0A067QPR6 E0VYN6 A0A1B0DN89 A0A1B6DNU0 A0A182JRJ7 T1IUT3 T1IDN1 A0A3S3PZD6 A0A1I8Q2Q8 K1RB06 A0A1J1INU8 A0A2J7R5X9 E2B718 K1Q1E6 A0A2J7R5X6 U4U1C6 N6U6B5 U4UTH7 K7IY54 A0A1I8MTR3 A0A2W1BM30 A0A151IPS5 A0A026X1E7 A0A087T143 A0A2P8YAH3 A0A195DZ05 A0A151X0G3 A0A2L2YJ02 A0A195EWS7 F4W5Q6 A0A195BTB1 A0A1B0AJA1 A0A1A9V2J2 E1ZY92 A0A1B0GDU8 A0A2A3ESR3 X1WJK4 A0A1A9X438 A0A3S3NU32 A0A0L8HAX3 A0A1I8Q2R0 V4BRA1 A0A1I8Q2N3
A0A2H1VJH0 A0A2A4K8N2 D6WJE5 A0A0T6AXB0 A0A336LZA2 A0A194Q763 A0A2R7WC52 B4LZX9 A0A0M9A6K9 A0A182VQ70 A0A0L7QSC4 A0A1Y1MZ42 A0A182RPN3 A0A182QRZ4 A0A087ZVH2 A0A0A9XK59 A0A182FNS2 W5JNU8 A0A182N4Q8 Q7QAP4 A0A154P2D5 A0A182HZC6 A0A182WVR2 A0A182KXI8 A0A182VB63 A0A182P929 A0A310SK20 A0A182IU05 A0A182TG03 A0A182Y7C0 A0A182ME46 A0A084VHA2 B4K5U0 B4GLC4 Q294A6 W8AR20 B0XIU0 A0A1S4G777 A0A1Q3F0A8 B4JGD9 A0A0A9XQ18 A0A1B6K7B3 A0A2H1VSZ8 Q0IEE1 A0A1B0ET90 Q95TI3 Q9VF91 A0A3B0KEN1 A0A067QPR6 E0VYN6 A0A1B0DN89 A0A1B6DNU0 A0A182JRJ7 T1IUT3 T1IDN1 A0A3S3PZD6 A0A1I8Q2Q8 K1RB06 A0A1J1INU8 A0A2J7R5X9 E2B718 K1Q1E6 A0A2J7R5X6 U4U1C6 N6U6B5 U4UTH7 K7IY54 A0A1I8MTR3 A0A2W1BM30 A0A151IPS5 A0A026X1E7 A0A087T143 A0A2P8YAH3 A0A195DZ05 A0A151X0G3 A0A2L2YJ02 A0A195EWS7 F4W5Q6 A0A195BTB1 A0A1B0AJA1 A0A1A9V2J2 E1ZY92 A0A1B0GDU8 A0A2A3ESR3 X1WJK4 A0A1A9X438 A0A3S3NU32 A0A0L8HAX3 A0A1I8Q2R0 V4BRA1 A0A1I8Q2N3
Ontologies
GO
PANTHER
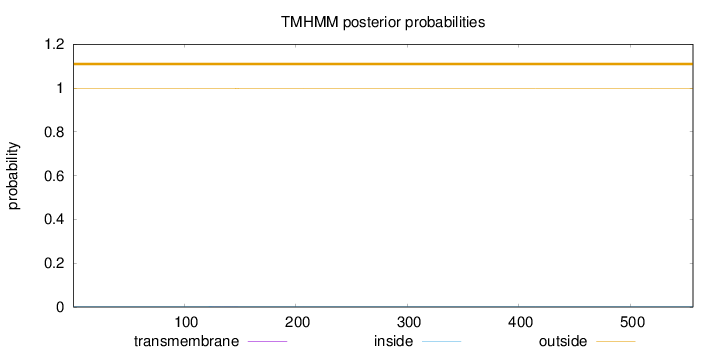
Topology
Length:
556
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02122
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00216
outside
1 - 556
Population Genetic Test Statistics
Pi
5.54798
Theta
16.829526
Tajima's D
-1.220064
CLR
0.304184
CSRT
0.095645217739113
Interpretation
Uncertain
