Gene
KWMTBOMO14291
Pre Gene Modal
BGIBMGA013424
Annotation
PREDICTED:_rho_GTPase-activating_protein_190_isoform_X1_[Bombyx_mori]
Full name
Rho GTPase-activating protein 190
Alternative Name
Rho GTPase-activating protein of 190 kDa
Location in the cell
Cytoplasmic Reliability : 1.352 Mitochondrial Reliability : 1.654
Sequence
CDS
ATGGCGAGGAAAAGGAACTTGGTGAAGTGCATAGAGTTCATAGAGCGCGAAGGGCTGGGCTCTGAGGGACTCTACCGCGTGCCGGGGAACAGGGCTCACGTCGATATGCTCTTCACCAAGTTCTACGAAGATCCGAATATAGATTTAGATTCGCTAGACATACCCGTGAATGCTGTCGCCACTGCGCTCAAGGATTTCTTCTCGAAGAAATTACCGCCATTACTCGATGAGGCGAGCATGGCGCAGTTGGAAGATATCGCGAAAGTGCACGTTGTGGGTACAGCGATGCGCGGCTGCATGGCGGGCGGCGTGGAACTCAAGGACCGCAGCTGGCGGCTGCTGGCCCTGCGCGCGCTGCTGCACTCCGCGCTCACGCCGCTCGCGCGCGCCACGCTCGACTACCTGCTGCACCACTTCGCCAGAGTCGCCGACAACTCATACCTGAACCCGATGTTGCGGTAG
Protein
MARKRNLVKCIEFIEREGLGSEGLYRVPGNRAHVDMLFTKFYEDPNIDLDSLDIPVNAVATALKDFFSKKLPPLLDEASMAQLEDIAKVHVVGTAMRGCMAGGVELKDRSWRLLALRALLHSALTPLARATLDYLLHHFARVADNSYLNPMLR
Summary
Description
GTPase-activating protein (GAP) for RhoA/Rho1 that plays an essential role in the stability of dorsal branches of mushroom body (MB) neurons. The MB neurons are the center for olfactory learning and memory. Acts by converting RhoA/Rho1 to an inactive GDP-bound state, leading to repress the RhoA/Rho1-Drok-MRLC signaling pathway thereby maintaining axon branch stability.
Keywords
Alternative splicing
Coiled coil
Complete proteome
Developmental protein
GTPase activation
Phosphoprotein
Reference proteome
Repeat
Feature
chain Rho GTPase-activating protein 190
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JV61
A0A3S2TRZ6
A0A2A4JSL4
A0A2H1V6C9
A0A194PVW5
W8BQ35
+ More
B4N201 A0A1B0FY42 A0A1Y1KVX8 A0A1W4XDQ3 B4R6L8 A0A0R1EBZ8 A0A0R1EBX3 A0A0Q5T3Z0 M9PJQ3 A0A0Q9WPF4 A0A0Q9WE01 A0A0Q9WPQ1 M9PHV3 B4IF32 B4MAZ2 B4Q2I7 Q9VX32-2 M9PHG0 A0A0R1EBJ3 A0A0Q9WQX0 A0A0Q5T3P9 B3NWW3 A0A0C5KMR0 Q9VX32 A0A0Q5T3R5 A0A0R1EI73 M9PHY4 A0A0R1EBR4 A0A0Q9WDF1 A0A0Q5T6H5 A0A0Q5TAE3 A0A0Q9WNN5 B4JNF5 A0A0M3QZ95 A0A1Y1KQ68 A0A1Y1KQ67 A0A0P8ZSL1 B3MY00 A0A0P8Y7S2 W8ARY8 W8AZE6 A0A1W4V836 A0A1W4V906 A0A1W4UVB7 A0A1W4V910 A0A1W4UVQ2 A0A1W4V842 W8BEI6 A0A139WPT7 A0A139WPM0 D6W848 A0A139WPL6 A0A139WPB3 A0A139WPB0 A0A2A3EBP9 A0A0R3P3J5 A0A0Q9XES7 A0A0Q9XE93 A0A0Q9XNN6 A0A0Q9XE78 A0A0Q9XPI0 A0A1Y1KSX7 A0A0R3NY69 A0A0R3NXX8 B4L5E1 A0A0Q9XE92 A0A0Q9XSK8 A0A0Q9XQR9 A0A0R3P485 B5DKM2 A0A0R3P3N5 A0A0R3P2Z7 A0A0R3NXW7 B4H9W9 A0A087ZTU0 A0A3B0J6L2 A0A3B0JWS2 A0A3B0J7L7 A0A3B0JBC8 A0A0L7RBB4 N6TE97 A0A1Y1KTM3 A0A034W094 A0A0K8VJ38 A0A034W1V2 A0A0K8VIK8 A0A0C9QV64 A0A0C9RH51 A0A0C9RL47 A0A3B0JCI9 E2A9N1 A0A151X3G2 A0A195ERU0
B4N201 A0A1B0FY42 A0A1Y1KVX8 A0A1W4XDQ3 B4R6L8 A0A0R1EBZ8 A0A0R1EBX3 A0A0Q5T3Z0 M9PJQ3 A0A0Q9WPF4 A0A0Q9WE01 A0A0Q9WPQ1 M9PHV3 B4IF32 B4MAZ2 B4Q2I7 Q9VX32-2 M9PHG0 A0A0R1EBJ3 A0A0Q9WQX0 A0A0Q5T3P9 B3NWW3 A0A0C5KMR0 Q9VX32 A0A0Q5T3R5 A0A0R1EI73 M9PHY4 A0A0R1EBR4 A0A0Q9WDF1 A0A0Q5T6H5 A0A0Q5TAE3 A0A0Q9WNN5 B4JNF5 A0A0M3QZ95 A0A1Y1KQ68 A0A1Y1KQ67 A0A0P8ZSL1 B3MY00 A0A0P8Y7S2 W8ARY8 W8AZE6 A0A1W4V836 A0A1W4V906 A0A1W4UVB7 A0A1W4V910 A0A1W4UVQ2 A0A1W4V842 W8BEI6 A0A139WPT7 A0A139WPM0 D6W848 A0A139WPL6 A0A139WPB3 A0A139WPB0 A0A2A3EBP9 A0A0R3P3J5 A0A0Q9XES7 A0A0Q9XE93 A0A0Q9XNN6 A0A0Q9XE78 A0A0Q9XPI0 A0A1Y1KSX7 A0A0R3NY69 A0A0R3NXX8 B4L5E1 A0A0Q9XE92 A0A0Q9XSK8 A0A0Q9XQR9 A0A0R3P485 B5DKM2 A0A0R3P3N5 A0A0R3P2Z7 A0A0R3NXW7 B4H9W9 A0A087ZTU0 A0A3B0J6L2 A0A3B0JWS2 A0A3B0J7L7 A0A3B0JBC8 A0A0L7RBB4 N6TE97 A0A1Y1KTM3 A0A034W094 A0A0K8VJ38 A0A034W1V2 A0A0K8VIK8 A0A0C9QV64 A0A0C9RH51 A0A0C9RL47 A0A3B0JCI9 E2A9N1 A0A151X3G2 A0A195ERU0
Pubmed
EMBL
BABH01026464
RSAL01000012
RVE53521.1
NWSH01000653
PCG75001.1
ODYU01000720
+ More
SOQ35952.1 KQ459590 KPI97467.1 GAMC01014946 JAB91609.1 CH963925 EDW78390.2 AJVK01005168 GEZM01076912 JAV63556.1 CM000366 EDX18217.1 CM000162 KRK06857.1 KRK06858.1 KRK06861.1 KRK06862.1 CH954180 KQS29997.1 AE014298 AGB95501.1 CH940655 KRF82665.1 KRF82669.1 KRF82670.1 KRF82666.1 KRF82667.1 AGB95500.1 CH480832 EDW46286.1 EDW66401.2 EDX02628.1 KRK06860.1 AF387518 AY121666 AGB95498.1 KRK06863.1 KRK06864.1 KRF82672.1 KQS29995.1 KQS29999.1 KQS30003.1 EDV46510.1 KQS29996.1 BT150459 AJP62081.1 KQS29994.1 KQS30002.1 KRK06866.1 AGB95499.1 KRK06859.1 KRK06865.1 KRF82668.1 KQS30000.1 KQS29998.1 KQS30001.1 KRF82671.1 CH916371 EDV92248.1 CP012528 ALC49033.1 GEZM01076914 JAV63552.1 GEZM01076910 JAV63559.1 CH902630 KPU77485.1 EDV38615.2 KPU77486.1 GAMC01014945 JAB91610.1 GAMC01014943 JAB91612.1 GAMC01014944 JAB91611.1 KQ971307 KYB29835.1 KYB29836.1 EFA10998.2 KYB29837.1 KYB29838.1 KYB29839.1 KZ288291 PBC29203.1 CH379063 KRT05920.1 KRT05927.1 CH933811 KRG06893.1 KRG06895.1 KRG06900.1 KRG06896.1 KRG06894.1 GEZM01076915 JAV63551.1 KRT05924.1 KRT05921.1 EDW06400.2 KRG06892.1 KRG06899.1 KRG06897.1 KRT05926.1 EDY72072.1 KRT05923.1 KRT05925.1 KRT05922.1 CH479232 EDW36626.1 OUUW01000003 SPP77804.1 SPP77806.1 SPP77805.1 SPP77803.1 KQ414617 KOC68212.1 APGK01032712 KB740848 KB632411 ENN78679.1 ERL95234.1 GEZM01076909 JAV63560.1 GAKP01010838 JAC48114.1 GDHF01013437 GDHF01003775 JAI38877.1 JAI48539.1 GAKP01010837 JAC48115.1 GDHF01017657 GDHF01013929 JAI34657.1 JAI38385.1 GBYB01007639 JAG77406.1 GBYB01007640 JAG77407.1 GBYB01007641 JAG77408.1 SPP77802.1 GL437918 EFN69833.1 KQ982562 KYQ54932.1 KQ981993 KYN30896.1
SOQ35952.1 KQ459590 KPI97467.1 GAMC01014946 JAB91609.1 CH963925 EDW78390.2 AJVK01005168 GEZM01076912 JAV63556.1 CM000366 EDX18217.1 CM000162 KRK06857.1 KRK06858.1 KRK06861.1 KRK06862.1 CH954180 KQS29997.1 AE014298 AGB95501.1 CH940655 KRF82665.1 KRF82669.1 KRF82670.1 KRF82666.1 KRF82667.1 AGB95500.1 CH480832 EDW46286.1 EDW66401.2 EDX02628.1 KRK06860.1 AF387518 AY121666 AGB95498.1 KRK06863.1 KRK06864.1 KRF82672.1 KQS29995.1 KQS29999.1 KQS30003.1 EDV46510.1 KQS29996.1 BT150459 AJP62081.1 KQS29994.1 KQS30002.1 KRK06866.1 AGB95499.1 KRK06859.1 KRK06865.1 KRF82668.1 KQS30000.1 KQS29998.1 KQS30001.1 KRF82671.1 CH916371 EDV92248.1 CP012528 ALC49033.1 GEZM01076914 JAV63552.1 GEZM01076910 JAV63559.1 CH902630 KPU77485.1 EDV38615.2 KPU77486.1 GAMC01014945 JAB91610.1 GAMC01014943 JAB91612.1 GAMC01014944 JAB91611.1 KQ971307 KYB29835.1 KYB29836.1 EFA10998.2 KYB29837.1 KYB29838.1 KYB29839.1 KZ288291 PBC29203.1 CH379063 KRT05920.1 KRT05927.1 CH933811 KRG06893.1 KRG06895.1 KRG06900.1 KRG06896.1 KRG06894.1 GEZM01076915 JAV63551.1 KRT05924.1 KRT05921.1 EDW06400.2 KRG06892.1 KRG06899.1 KRG06897.1 KRT05926.1 EDY72072.1 KRT05923.1 KRT05925.1 KRT05922.1 CH479232 EDW36626.1 OUUW01000003 SPP77804.1 SPP77806.1 SPP77805.1 SPP77803.1 KQ414617 KOC68212.1 APGK01032712 KB740848 KB632411 ENN78679.1 ERL95234.1 GEZM01076909 JAV63560.1 GAKP01010838 JAC48114.1 GDHF01013437 GDHF01003775 JAI38877.1 JAI48539.1 GAKP01010837 JAC48115.1 GDHF01017657 GDHF01013929 JAI34657.1 JAI38385.1 GBYB01007639 JAG77406.1 GBYB01007640 JAG77407.1 GBYB01007641 JAG77408.1 SPP77802.1 GL437918 EFN69833.1 KQ982562 KYQ54932.1 KQ981993 KYN30896.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007798
UP000092462
+ More
UP000192223 UP000000304 UP000002282 UP000008711 UP000000803 UP000008792 UP000001292 UP000001070 UP000092553 UP000007801 UP000192221 UP000007266 UP000242457 UP000001819 UP000009192 UP000008744 UP000005203 UP000268350 UP000053825 UP000019118 UP000030742 UP000000311 UP000075809 UP000078541
UP000192223 UP000000304 UP000002282 UP000008711 UP000000803 UP000008792 UP000001292 UP000001070 UP000092553 UP000007801 UP000192221 UP000007266 UP000242457 UP000001819 UP000009192 UP000008744 UP000005203 UP000268350 UP000053825 UP000019118 UP000030742 UP000000311 UP000075809 UP000078541
Interpro
IPR008936
Rho_GTPase_activation_prot
+ More
IPR000198 RhoGAP_dom
IPR032835 RhoGAP-FF1
IPR027417 P-loop_NTPase
IPR039007 pG1
IPR039006 RhoGAP_pG2
IPR001806 Small_GTPase
IPR002713 FF_domain
IPR036517 FF_domain_sf
IPR002401 Cyt_P450_E_grp-I
IPR017972 Cyt_P450_CS
IPR001128 Cyt_P450
IPR036396 Cyt_P450_sf
IPR000198 RhoGAP_dom
IPR032835 RhoGAP-FF1
IPR027417 P-loop_NTPase
IPR039007 pG1
IPR039006 RhoGAP_pG2
IPR001806 Small_GTPase
IPR002713 FF_domain
IPR036517 FF_domain_sf
IPR002401 Cyt_P450_E_grp-I
IPR017972 Cyt_P450_CS
IPR001128 Cyt_P450
IPR036396 Cyt_P450_sf
Gene 3D
ProteinModelPortal
H9JV61
A0A3S2TRZ6
A0A2A4JSL4
A0A2H1V6C9
A0A194PVW5
W8BQ35
+ More
B4N201 A0A1B0FY42 A0A1Y1KVX8 A0A1W4XDQ3 B4R6L8 A0A0R1EBZ8 A0A0R1EBX3 A0A0Q5T3Z0 M9PJQ3 A0A0Q9WPF4 A0A0Q9WE01 A0A0Q9WPQ1 M9PHV3 B4IF32 B4MAZ2 B4Q2I7 Q9VX32-2 M9PHG0 A0A0R1EBJ3 A0A0Q9WQX0 A0A0Q5T3P9 B3NWW3 A0A0C5KMR0 Q9VX32 A0A0Q5T3R5 A0A0R1EI73 M9PHY4 A0A0R1EBR4 A0A0Q9WDF1 A0A0Q5T6H5 A0A0Q5TAE3 A0A0Q9WNN5 B4JNF5 A0A0M3QZ95 A0A1Y1KQ68 A0A1Y1KQ67 A0A0P8ZSL1 B3MY00 A0A0P8Y7S2 W8ARY8 W8AZE6 A0A1W4V836 A0A1W4V906 A0A1W4UVB7 A0A1W4V910 A0A1W4UVQ2 A0A1W4V842 W8BEI6 A0A139WPT7 A0A139WPM0 D6W848 A0A139WPL6 A0A139WPB3 A0A139WPB0 A0A2A3EBP9 A0A0R3P3J5 A0A0Q9XES7 A0A0Q9XE93 A0A0Q9XNN6 A0A0Q9XE78 A0A0Q9XPI0 A0A1Y1KSX7 A0A0R3NY69 A0A0R3NXX8 B4L5E1 A0A0Q9XE92 A0A0Q9XSK8 A0A0Q9XQR9 A0A0R3P485 B5DKM2 A0A0R3P3N5 A0A0R3P2Z7 A0A0R3NXW7 B4H9W9 A0A087ZTU0 A0A3B0J6L2 A0A3B0JWS2 A0A3B0J7L7 A0A3B0JBC8 A0A0L7RBB4 N6TE97 A0A1Y1KTM3 A0A034W094 A0A0K8VJ38 A0A034W1V2 A0A0K8VIK8 A0A0C9QV64 A0A0C9RH51 A0A0C9RL47 A0A3B0JCI9 E2A9N1 A0A151X3G2 A0A195ERU0
B4N201 A0A1B0FY42 A0A1Y1KVX8 A0A1W4XDQ3 B4R6L8 A0A0R1EBZ8 A0A0R1EBX3 A0A0Q5T3Z0 M9PJQ3 A0A0Q9WPF4 A0A0Q9WE01 A0A0Q9WPQ1 M9PHV3 B4IF32 B4MAZ2 B4Q2I7 Q9VX32-2 M9PHG0 A0A0R1EBJ3 A0A0Q9WQX0 A0A0Q5T3P9 B3NWW3 A0A0C5KMR0 Q9VX32 A0A0Q5T3R5 A0A0R1EI73 M9PHY4 A0A0R1EBR4 A0A0Q9WDF1 A0A0Q5T6H5 A0A0Q5TAE3 A0A0Q9WNN5 B4JNF5 A0A0M3QZ95 A0A1Y1KQ68 A0A1Y1KQ67 A0A0P8ZSL1 B3MY00 A0A0P8Y7S2 W8ARY8 W8AZE6 A0A1W4V836 A0A1W4V906 A0A1W4UVB7 A0A1W4V910 A0A1W4UVQ2 A0A1W4V842 W8BEI6 A0A139WPT7 A0A139WPM0 D6W848 A0A139WPL6 A0A139WPB3 A0A139WPB0 A0A2A3EBP9 A0A0R3P3J5 A0A0Q9XES7 A0A0Q9XE93 A0A0Q9XNN6 A0A0Q9XE78 A0A0Q9XPI0 A0A1Y1KSX7 A0A0R3NY69 A0A0R3NXX8 B4L5E1 A0A0Q9XE92 A0A0Q9XSK8 A0A0Q9XQR9 A0A0R3P485 B5DKM2 A0A0R3P3N5 A0A0R3P2Z7 A0A0R3NXW7 B4H9W9 A0A087ZTU0 A0A3B0J6L2 A0A3B0JWS2 A0A3B0J7L7 A0A3B0JBC8 A0A0L7RBB4 N6TE97 A0A1Y1KTM3 A0A034W094 A0A0K8VJ38 A0A034W1V2 A0A0K8VIK8 A0A0C9QV64 A0A0C9RH51 A0A0C9RL47 A0A3B0JCI9 E2A9N1 A0A151X3G2 A0A195ERU0
PDB
2EE5
E-value=7.24534e-14,
Score=182
Ontologies
GO
Topology
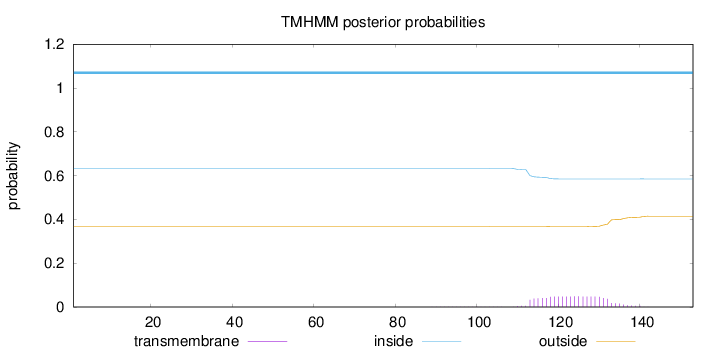
Length:
153
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.99552
Exp number, first 60 AAs:
0
Total prob of N-in:
0.63265
inside
1 - 153
Population Genetic Test Statistics
Pi
346.071863
Theta
132.034422
Tajima's D
5.075629
CLR
0.116471
CSRT
0.999950002499875
Interpretation
Uncertain
