Gene
KWMTBOMO14290 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008174
Annotation
insulin-related_peptide_binding_protein_precursor_[Bombyx_mori]
Full name
Neural/ectodermal development factor IMP-L2
Location in the cell
Extracellular Reliability : 2.443
Sequence
CDS
ATGCATTTAGTTCTGCTATTCACCGTCGCGGCGCTGCTGGGCTCGTGCCAGTCCGCCCATTTGAATAAACACATTAAGCTGCTTTCGGACATCGATAACAGTATTGAGAATGGTGTGCAAGCAAAATCTGATGGATCTCATAAATATTTATCGATCACGCAAGGCCCCCTACCGTCGTACGCACATACTCCCGGAACCACTATTGAGTTGACCTGCGAAGCTGCCGGATCTCCAGCACCATCAGTACACTGGTTCAAGAACGACTCTCCAGTCTACGAGTACGACGTCGAGTCCAACGAATTAATCGACTCAAGCCCGACATCTATTGCGAGAATTTCCTCAACTCTCATAGTGACGCGTACGACCTCTCAGGATGTGTACACCTGCCTCGCCACCACGAGCTCCAAGACTGCGCGAGCCAGCACTGTGGTCTACAATACAGATAGCGCTACTGAGCTCTCGGAACGCGCTAAGCTGTTCCCGCTGAAGCCTCGCATCGTGGTCTCCTATAGCACCTACGTGGACAACATCGGCAACAGGGTGGTGCTCCCGTGCCGCGTCAAGGGACACCCCAAGCCCAAGATCACCTGGTTCAACGGACAGAATGTGCCCATTGAAAAGAACCCGCGCATGAAGGTGCTTCGCTCGGGCGAGCTGGTCATATCCTCCCTCCTCTGGAGCGACATGGACGAGTACACTTGCCAAGCCGAAAACGCTTTCGGCTCGGAGAAGGCTAAAACATTCGTCTACCCCGCTAAAGTAAGTTTCTCAACACTATTCTTTCTATTGAATTAA
Protein
MHLVLLFTVAALLGSCQSAHLNKHIKLLSDIDNSIENGVQAKSDGSHKYLSITQGPLPSYAHTPGTTIELTCEAAGSPAPSVHWFKNDSPVYEYDVESNELIDSSPTSIARISSTLIVTRTTSQDVYTCLATTSSKTARASTVVYNTDSATELSERAKLFPLKPRIVVSYSTYVDNIGNRVVLPCRVKGHPKPKITWFNGQNVPIEKNPRMKVLRSGELVISSLLWSDMDEYTCQAENAFGSEKAKTFVYPAKVSFSTLFFLLN
Summary
Keywords
3D-structure
Alternative splicing
Cell adhesion
Complete proteome
Developmental protein
Disulfide bond
Immunoglobulin domain
Reference proteome
Repeat
Secreted
Signal
Feature
chain Neural/ectodermal development factor IMP-L2
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JF77
Q1HPN0
A0A2A4K9A1
A0A1E1WQL1
A0A2H1WEL3
A0A1E1W4I7
+ More
A0A3S2M383 S4PH80 A0A194QCP9 A0A212FK32 A0A0N1IGP5 A0A2W1BR81 Q9NGZ0 A0A2A4J7R6 A0A2H1WEZ6 A0A2W1BU10 A0A0Q9XR78 B4L9B7 I7B1E3 H9JF49 B4N3I2 A0A2M4BWB5 A0A2M4BVK8 A0A2M4BVS4 A0A182FQU3 A0A2M4A3E4 A0A2M3Z633 A0A2M4A3G9 A0A2M4A3L1 A0A2M4A3U8 A0A2M3Z5X2 W5JCF6 A0A2M3ZBT5 A0A2M4BQR2 A0A2M4BQZ3 A0A2M4A390 A0A2M3Z5M8 A0A2M3ZH72 A0A2M3ZHB9 A0A182T7Y4 A0A034VDX8 A0A034VCU9 T1DJH9 A0A182PSW2 A0A182Q760 B3M4X6 A0A182NSE2 A0A0P9C4H1 A0A182RYB9 A0A194Q6P9 A0A0A1XCB6 A0A0A1XL72 A0A0N1IHW9 A0A182HJ33 A0A0K8VFJ2 A0A182W2S8 A0A1W4W600 A0A1W4VTZ2 A0A182XVJ6 A0A182XE89 Q7QBJ8 A0A182MIJ8 A0A0M5IYQ6 A0A182UXE8 A0A1I8PUB4 A0A1I8PU90 A0A084VZY4 T1P9T8 A0A1I8MFY8 A0A182LC86 Q09024 Q3YMW0 Q09024-2 Q09024-3 M9PEL7 A0A182ILB4 B4PHS2 Q3YMV9 B4QQI2 A0A0R1E0W7 A0A0Q5UEF5 B3NC03 A0A0J9UED0 B4LBP5 A0A3B0JVS0 A0A3B0J6U1 B4J1G6 A0A0L0CHH7 G8FPZ5 W8BB38 W8AFL5 A0A0R3P972 Q29D65 A0A0Q9WUR3 A0A0Q9WSF5 B4HU27 A0A1A9WLJ4 A0A1B0FDT7 A0A1A9UG40 A0A1A9ZKF1
A0A3S2M383 S4PH80 A0A194QCP9 A0A212FK32 A0A0N1IGP5 A0A2W1BR81 Q9NGZ0 A0A2A4J7R6 A0A2H1WEZ6 A0A2W1BU10 A0A0Q9XR78 B4L9B7 I7B1E3 H9JF49 B4N3I2 A0A2M4BWB5 A0A2M4BVK8 A0A2M4BVS4 A0A182FQU3 A0A2M4A3E4 A0A2M3Z633 A0A2M4A3G9 A0A2M4A3L1 A0A2M4A3U8 A0A2M3Z5X2 W5JCF6 A0A2M3ZBT5 A0A2M4BQR2 A0A2M4BQZ3 A0A2M4A390 A0A2M3Z5M8 A0A2M3ZH72 A0A2M3ZHB9 A0A182T7Y4 A0A034VDX8 A0A034VCU9 T1DJH9 A0A182PSW2 A0A182Q760 B3M4X6 A0A182NSE2 A0A0P9C4H1 A0A182RYB9 A0A194Q6P9 A0A0A1XCB6 A0A0A1XL72 A0A0N1IHW9 A0A182HJ33 A0A0K8VFJ2 A0A182W2S8 A0A1W4W600 A0A1W4VTZ2 A0A182XVJ6 A0A182XE89 Q7QBJ8 A0A182MIJ8 A0A0M5IYQ6 A0A182UXE8 A0A1I8PUB4 A0A1I8PU90 A0A084VZY4 T1P9T8 A0A1I8MFY8 A0A182LC86 Q09024 Q3YMW0 Q09024-2 Q09024-3 M9PEL7 A0A182ILB4 B4PHS2 Q3YMV9 B4QQI2 A0A0R1E0W7 A0A0Q5UEF5 B3NC03 A0A0J9UED0 B4LBP5 A0A3B0JVS0 A0A3B0J6U1 B4J1G6 A0A0L0CHH7 G8FPZ5 W8BB38 W8AFL5 A0A0R3P972 Q29D65 A0A0Q9WUR3 A0A0Q9WSF5 B4HU27 A0A1A9WLJ4 A0A1B0FDT7 A0A1A9UG40 A0A1A9ZKF1
Pubmed
19121390
23622113
26354079
22118469
28756777
10748036
+ More
17994087 20920257 23761445 25348373 25830018 25244985 12364791 14747013 17210077 24438588 25315136 20966253 8306886 10731132 12537572 12537569 15917496 12537568 12537573 12537574 16110336 17569856 17569867 17550304 22936249 26108605 22401767 24495485 15632085 23185243
17994087 20920257 23761445 25348373 25830018 25244985 12364791 14747013 17210077 24438588 25315136 20966253 8306886 10731132 12537572 12537569 15917496 12537568 12537573 12537574 16110336 17569856 17569867 17550304 22936249 26108605 22401767 24495485 15632085 23185243
EMBL
BABH01026877
DQ443372
ABF51461.1
NWSH01000023
PCG80649.1
GDQN01008520
+ More
GDQN01001847 JAT82534.1 JAT89207.1 ODYU01008121 SOQ51457.1 GDQN01009180 JAT81874.1 RSAL01000055 RVE49949.1 GAIX01005890 JAA86670.1 KQ459386 KPJ01216.1 AGBW02008149 OWR54098.1 KQ460423 KPJ14843.1 KZ149968 PZC76125.1 AF236641 AAF61949.1 NWSH01002545 PCG68011.1 ODYU01008227 SOQ51639.1 PZC76126.1 CH933816 KRG07716.1 EDW17292.1 JX133233 AFO09967.1 BABH01026869 CH964095 EDW79187.2 GGFJ01008090 MBW57231.1 GGFJ01007891 MBW57032.1 GGFJ01007890 MBW57031.1 GGFK01002013 MBW35334.1 GGFM01003228 MBW23979.1 GGFK01002016 MBW35337.1 GGFK01001989 MBW35310.1 GGFK01001997 MBW35318.1 GGFM01003159 MBW23910.1 ADMH02001615 ETN61781.1 GGFM01005256 MBW26007.1 GGFJ01006284 MBW55425.1 GGFJ01006283 MBW55424.1 GGFK01001787 MBW35108.1 GGFM01003069 MBW23820.1 GGFM01007145 MBW27896.1 GGFM01007158 MBW27909.1 GAKP01018640 JAC40312.1 GAKP01018641 GAKP01018638 JAC40314.1 GAMD01001431 JAB00160.1 AXCN02000238 CH902618 EDV40550.1 KPU78626.1 KPJ01217.1 GBXI01006189 JAD08103.1 GBXI01002561 JAD11731.1 KPJ14842.1 APCN01002121 GDHF01015024 JAI37290.1 AAAB01008879 EAA08451.5 EGK96962.1 AXCM01001682 CP012525 ALC44018.1 ATLV01019039 KE525259 KFB43528.1 KA644713 AFP59342.1 L23066 AE014296 AY060476 BT012298 BT012299 DQ062766 AAY56639.1 AGB94104.1 CM000159 EDW93381.2 DQ062767 AAY56640.1 CM000363 CM002912 EDX09188.1 KMY97539.1 KRK01146.1 CH954178 KQS43569.1 EDV50891.1 KMY97540.1 CH940647 EDW68672.2 OUUW01000002 SPP77456.1 SPP77455.1 CH916366 EDV95857.1 JRES01000398 KNC31667.1 JN020646 AER29815.1 GAMC01019511 JAB87044.1 GAMC01019510 JAB87045.1 CH379070 KRT09043.1 EAL30549.3 KRT09042.1 KRF83968.1 KRF83967.1 CH480817 EDW50448.1 CCAG010009000 CCAG010009001
GDQN01001847 JAT82534.1 JAT89207.1 ODYU01008121 SOQ51457.1 GDQN01009180 JAT81874.1 RSAL01000055 RVE49949.1 GAIX01005890 JAA86670.1 KQ459386 KPJ01216.1 AGBW02008149 OWR54098.1 KQ460423 KPJ14843.1 KZ149968 PZC76125.1 AF236641 AAF61949.1 NWSH01002545 PCG68011.1 ODYU01008227 SOQ51639.1 PZC76126.1 CH933816 KRG07716.1 EDW17292.1 JX133233 AFO09967.1 BABH01026869 CH964095 EDW79187.2 GGFJ01008090 MBW57231.1 GGFJ01007891 MBW57032.1 GGFJ01007890 MBW57031.1 GGFK01002013 MBW35334.1 GGFM01003228 MBW23979.1 GGFK01002016 MBW35337.1 GGFK01001989 MBW35310.1 GGFK01001997 MBW35318.1 GGFM01003159 MBW23910.1 ADMH02001615 ETN61781.1 GGFM01005256 MBW26007.1 GGFJ01006284 MBW55425.1 GGFJ01006283 MBW55424.1 GGFK01001787 MBW35108.1 GGFM01003069 MBW23820.1 GGFM01007145 MBW27896.1 GGFM01007158 MBW27909.1 GAKP01018640 JAC40312.1 GAKP01018641 GAKP01018638 JAC40314.1 GAMD01001431 JAB00160.1 AXCN02000238 CH902618 EDV40550.1 KPU78626.1 KPJ01217.1 GBXI01006189 JAD08103.1 GBXI01002561 JAD11731.1 KPJ14842.1 APCN01002121 GDHF01015024 JAI37290.1 AAAB01008879 EAA08451.5 EGK96962.1 AXCM01001682 CP012525 ALC44018.1 ATLV01019039 KE525259 KFB43528.1 KA644713 AFP59342.1 L23066 AE014296 AY060476 BT012298 BT012299 DQ062766 AAY56639.1 AGB94104.1 CM000159 EDW93381.2 DQ062767 AAY56640.1 CM000363 CM002912 EDX09188.1 KMY97539.1 KRK01146.1 CH954178 KQS43569.1 EDV50891.1 KMY97540.1 CH940647 EDW68672.2 OUUW01000002 SPP77456.1 SPP77455.1 CH916366 EDV95857.1 JRES01000398 KNC31667.1 JN020646 AER29815.1 GAMC01019511 JAB87044.1 GAMC01019510 JAB87045.1 CH379070 KRT09043.1 EAL30549.3 KRT09042.1 KRF83968.1 KRF83967.1 CH480817 EDW50448.1 CCAG010009000 CCAG010009001
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
+ More
UP000009192 UP000007798 UP000069272 UP000000673 UP000075901 UP000075885 UP000075886 UP000007801 UP000075884 UP000075900 UP000075840 UP000075920 UP000192221 UP000076408 UP000076407 UP000007062 UP000075883 UP000092553 UP000075903 UP000095300 UP000030765 UP000095301 UP000075882 UP000000803 UP000075880 UP000002282 UP000000304 UP000008711 UP000008792 UP000268350 UP000001070 UP000037069 UP000001819 UP000001292 UP000091820 UP000092444 UP000078200 UP000092445
UP000009192 UP000007798 UP000069272 UP000000673 UP000075901 UP000075885 UP000075886 UP000007801 UP000075884 UP000075900 UP000075840 UP000075920 UP000192221 UP000076408 UP000076407 UP000007062 UP000075883 UP000092553 UP000075903 UP000095300 UP000030765 UP000095301 UP000075882 UP000000803 UP000075880 UP000002282 UP000000304 UP000008711 UP000008792 UP000268350 UP000001070 UP000037069 UP000001819 UP000001292 UP000091820 UP000092444 UP000078200 UP000092445
Pfam
PF07679 I-set
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
H9JF77
Q1HPN0
A0A2A4K9A1
A0A1E1WQL1
A0A2H1WEL3
A0A1E1W4I7
+ More
A0A3S2M383 S4PH80 A0A194QCP9 A0A212FK32 A0A0N1IGP5 A0A2W1BR81 Q9NGZ0 A0A2A4J7R6 A0A2H1WEZ6 A0A2W1BU10 A0A0Q9XR78 B4L9B7 I7B1E3 H9JF49 B4N3I2 A0A2M4BWB5 A0A2M4BVK8 A0A2M4BVS4 A0A182FQU3 A0A2M4A3E4 A0A2M3Z633 A0A2M4A3G9 A0A2M4A3L1 A0A2M4A3U8 A0A2M3Z5X2 W5JCF6 A0A2M3ZBT5 A0A2M4BQR2 A0A2M4BQZ3 A0A2M4A390 A0A2M3Z5M8 A0A2M3ZH72 A0A2M3ZHB9 A0A182T7Y4 A0A034VDX8 A0A034VCU9 T1DJH9 A0A182PSW2 A0A182Q760 B3M4X6 A0A182NSE2 A0A0P9C4H1 A0A182RYB9 A0A194Q6P9 A0A0A1XCB6 A0A0A1XL72 A0A0N1IHW9 A0A182HJ33 A0A0K8VFJ2 A0A182W2S8 A0A1W4W600 A0A1W4VTZ2 A0A182XVJ6 A0A182XE89 Q7QBJ8 A0A182MIJ8 A0A0M5IYQ6 A0A182UXE8 A0A1I8PUB4 A0A1I8PU90 A0A084VZY4 T1P9T8 A0A1I8MFY8 A0A182LC86 Q09024 Q3YMW0 Q09024-2 Q09024-3 M9PEL7 A0A182ILB4 B4PHS2 Q3YMV9 B4QQI2 A0A0R1E0W7 A0A0Q5UEF5 B3NC03 A0A0J9UED0 B4LBP5 A0A3B0JVS0 A0A3B0J6U1 B4J1G6 A0A0L0CHH7 G8FPZ5 W8BB38 W8AFL5 A0A0R3P972 Q29D65 A0A0Q9WUR3 A0A0Q9WSF5 B4HU27 A0A1A9WLJ4 A0A1B0FDT7 A0A1A9UG40 A0A1A9ZKF1
A0A3S2M383 S4PH80 A0A194QCP9 A0A212FK32 A0A0N1IGP5 A0A2W1BR81 Q9NGZ0 A0A2A4J7R6 A0A2H1WEZ6 A0A2W1BU10 A0A0Q9XR78 B4L9B7 I7B1E3 H9JF49 B4N3I2 A0A2M4BWB5 A0A2M4BVK8 A0A2M4BVS4 A0A182FQU3 A0A2M4A3E4 A0A2M3Z633 A0A2M4A3G9 A0A2M4A3L1 A0A2M4A3U8 A0A2M3Z5X2 W5JCF6 A0A2M3ZBT5 A0A2M4BQR2 A0A2M4BQZ3 A0A2M4A390 A0A2M3Z5M8 A0A2M3ZH72 A0A2M3ZHB9 A0A182T7Y4 A0A034VDX8 A0A034VCU9 T1DJH9 A0A182PSW2 A0A182Q760 B3M4X6 A0A182NSE2 A0A0P9C4H1 A0A182RYB9 A0A194Q6P9 A0A0A1XCB6 A0A0A1XL72 A0A0N1IHW9 A0A182HJ33 A0A0K8VFJ2 A0A182W2S8 A0A1W4W600 A0A1W4VTZ2 A0A182XVJ6 A0A182XE89 Q7QBJ8 A0A182MIJ8 A0A0M5IYQ6 A0A182UXE8 A0A1I8PUB4 A0A1I8PU90 A0A084VZY4 T1P9T8 A0A1I8MFY8 A0A182LC86 Q09024 Q3YMW0 Q09024-2 Q09024-3 M9PEL7 A0A182ILB4 B4PHS2 Q3YMV9 B4QQI2 A0A0R1E0W7 A0A0Q5UEF5 B3NC03 A0A0J9UED0 B4LBP5 A0A3B0JVS0 A0A3B0J6U1 B4J1G6 A0A0L0CHH7 G8FPZ5 W8BB38 W8AFL5 A0A0R3P972 Q29D65 A0A0Q9WUR3 A0A0Q9WSF5 B4HU27 A0A1A9WLJ4 A0A1B0FDT7 A0A1A9UG40 A0A1A9ZKF1
PDB
4CBP
E-value=1.0057e-34,
Score=365
Ontologies
GO
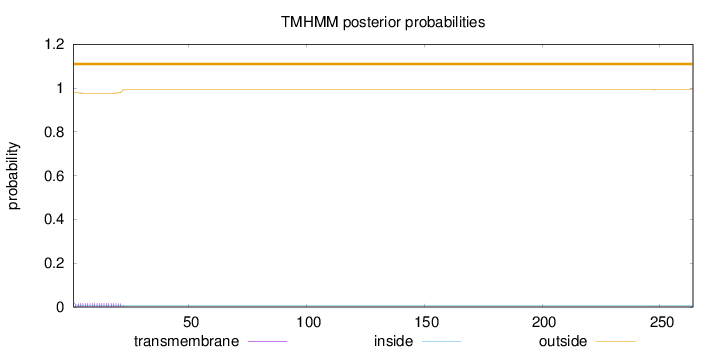
Topology
Subcellular location
SignalP
Position: 1 - 18,
Likelihood: 0.996970
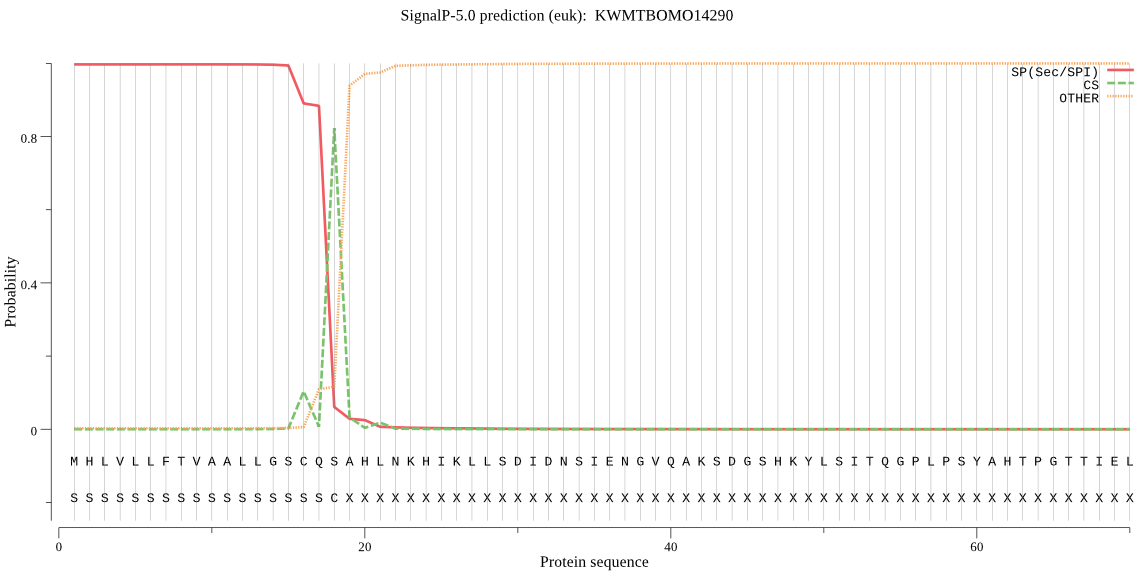
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.348850000000001
Exp number, first 60 AAs:
0.33403
Total prob of N-in:
0.02187
outside
1 - 264
Population Genetic Test Statistics
Pi
206.480986
Theta
159.705471
Tajima's D
1.054599
CLR
0.000105
CSRT
0.679366031698415
Interpretation
Uncertain
