Gene
KWMTBOMO14279
Pre Gene Modal
BGIBMGA008170
Annotation
PREDICTED:_EH_domain-binding_protein_1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.695
Sequence
CDS
ATGGCGTCAGTTTGGAAGCGATTGCAACGCGTCAACAAACGTGCGGCCAAGTTCCAGTACACCGCCTCGTATCACAGGGTCGACTTGGAGACCTCGCCCAAATGGAAACCTAATAAGTTAAGTGTAGTCTGGACGCGGAGATCTCGGAGAGTCATCAGCGAGCCTCTAGAATGGGAGCCAACTCTCAAGGACCCTTTGAAGGGATCGGTGATATACAATCTACCAGAGAATCACACCGTGGCGGTCACTCTCTTCAAGGATTCGCGCACTAATGAACTGGAGGACAAAGACTGGACTTTTGTACTGGAGGATGTGTCGTTGACTGGCAAGCGGCGTCGGCTGGCCGTGTGCTCCGTGAACATGCGCAAGTACGCGAGCCTGGAGTCGTCCCAGCGGCCGCTGCTGCTGCGGCTGGAGCCCACCACGCACAAGATTGTGTCCGCCTCCCTGTCGCTCACGTTACATTGCGTGCTGCTCAGAGAGGGACAGGCTACGGACGAGGACATGCAGTCGCTGGTCAGTCTGCTGTCGGTCAACAACAACTCCGACATCGCCGTGCTGGAGGACCTGGACGAGGACGACTACACGCTGTCCGATGACTCCACTCGCCAGATGTTGGACTTCCGTCAGCAAATGGAAGAGATGACCCAGAGCCTCACGAACAGCGACCTGGCGGACACCCCGTGCAGCGTGCAGAGCCTCAACCTCGACCTCACGCCGCAGCAGGCCCCGCCCGACTCGCCCACCACCCCGGTGCCCAGAGACGACAGACTCGATTTAAAAATAAAAATCGATTTACCCAAAATCGATTCCGTGACTTCCACCCCGAAAATCGATAAAACGGAATTCAAGCCTCTAGTATTGAAGCCCAGGACGATAACGAAAAGGAACCTTAAGCCTTTGGAGTTAAAGACGAATCTCAACAGCATCGGGCTCGAGAAGGACGATGAGAACAGCTACGACAGCGACCAGACGCCGACCGCCGACGTGGACAAACTGCTCCCGGGGAGGTTCCACAGAGACAAGACGCCGGTCCAAGACCTGCTGGAGTGGTGCCAGGCGGTGACCCGCGACTACAAGACCTGCCGGGTCACGAACCTCACCACCAGCTTCAGGTCCGGGCTGGCCTTCTGCGCCATCATACACCACTTTCGACCAGACCTCATCGACCTGTCGGCGCTGGCGGGCGAGGGGGCGGAGGCGCACGTGCGGGCGGCGCTGGAGGCCGGGGCCGCGCTGGGCATCCCGCAGGTGCTGGCCGTGGCCGACGTGTGCGCGCGTCCGGCGCCCGACCGGCTAGCCATCATGACGTACCTGTTCCAACTGCGGGCGCACTTCACGGGCGGCGAGCTGCAGGTGGAGCAACTGGGTAACGACGAGACGGAGTCGTCGTACATGGTGGGGCGCCACGACACGGACGAGTCGCTGCCCCCCGAGCTGTTCAGCCGCGAGATCAGCACGCGCCGCTCCAGGACCGACCGCGCACGACCCAAGTCTGAAGAACTAGAAGACAAGAAGAATAGAGTGGAGGAGCGCGGCAGTCCGGAGCGCGGCGGGGTGCGGGCGCTGCTGGGCCGCGTGCTGTCCCCGCAGCAGCAGCCGCCCACCTGGTACACGGACCAGCCGCTACCCGCTCGGCCGGAAAAGCTGATGACCCGCAAGGAACTGACCGATCCTTTCGGGTCCGACGAGGAGGACGAGCCGGGACCCCGGGGGACGCCCGAGCCACGAAGCAACGGAAAACTGGAAACAGCGGTGGAGCCCGCGCCGGACCCGCCGCGCATGGAGCCGGCGGCGACGGACCCCCTCACCCCCCCGGCGCCCCCCGCCGAGCCTAAACCGGAAACGCCGGTCGCAGAGCTGCCCAAGCCCACTCCGCAACTGTTGTCGCGGCACGATGAGCTCAAAGAGCGAGCGAGACAGCTGCTCGAGGCCACCAAGAGAGAGGCGAGAGAGAAAGAGAGGATCAGGCGACAGAACTCTAAGGAGGATAAAGAACTCAATGGCGTTAAGAATGGCGACTTGAGTCCGAAGAAGATAATGACGGAGGAAGAGAGACAAGTGATGCTGCGGGAGCGCGCGCGGAAGCTCATAGCCGACGCGAAAGCTGGACTGGTCAGTCCGAGTTCGCCGCTGTCTCCGTCCCGGCCGCCCCGCGCCGGCGCCGTGGAGATCATCAGCAAGCAGTCGGTGCTGGACGAGCTCGCTGCAGAGGACAGACAGAGCGAGGACGCGAAGAGCACCAGGAGTAACTCGTGGGGCAGTGACGAGGCGCGGCGGTCCCCCCTGCAGTCCTTCACCGCGCTCGTGGACAGCCTGTCGCCCGAGGACCACGCCGCGGATGAGGGCAAGGAGTCCGGCTCGTACATCCAGAGCGAGCTGGAAGCCTTGGAGCGAGAGCAGGCCGCCATCGACCTCAAGGCAGCCGCCCTAGAGAAGCAGTTACGCCACGTCATGGAGGCCGCGGACAGCACCGAGGAGGAGGACAAATTGATGTCGCAATGGTTCAACCTAGTCAATAAGAAGAACGCACTCCTGAGGAGGCAGATGCAGCTGAACATATTGGAGCAGGAGGAGGACCTGAGCCGCCGCTGCGAGCTGCTGGCGCGCGAGCTGCGCCTCTCCCTCGGCGTCGACGAGTGGAGGAAGACTCCCGGGCAGAAGAGGAGGGAGAGGCTGCTGCTGCAGGAGCTGCTGTCCGCGGTCAACGAGCGAGACCGCCTGGTCCAGGAGATGGACGAGCAGGAGAAAGCGATCGCGGACGACGACGCAATCGAGCGGAACCTGTCGCACGTCGAGATCCAAAGGAAGAACAACTGCATCCTGCAGTAG
Protein
MASVWKRLQRVNKRAAKFQYTASYHRVDLETSPKWKPNKLSVVWTRRSRRVISEPLEWEPTLKDPLKGSVIYNLPENHTVAVTLFKDSRTNELEDKDWTFVLEDVSLTGKRRRLAVCSVNMRKYASLESSQRPLLLRLEPTTHKIVSASLSLTLHCVLLREGQATDEDMQSLVSLLSVNNNSDIAVLEDLDEDDYTLSDDSTRQMLDFRQQMEEMTQSLTNSDLADTPCSVQSLNLDLTPQQAPPDSPTTPVPRDDRLDLKIKIDLPKIDSVTSTPKIDKTEFKPLVLKPRTITKRNLKPLELKTNLNSIGLEKDDENSYDSDQTPTADVDKLLPGRFHRDKTPVQDLLEWCQAVTRDYKTCRVTNLTTSFRSGLAFCAIIHHFRPDLIDLSALAGEGAEAHVRAALEAGAALGIPQVLAVADVCARPAPDRLAIMTYLFQLRAHFTGGELQVEQLGNDETESSYMVGRHDTDESLPPELFSREISTRRSRTDRARPKSEELEDKKNRVEERGSPERGGVRALLGRVLSPQQQPPTWYTDQPLPARPEKLMTRKELTDPFGSDEEDEPGPRGTPEPRSNGKLETAVEPAPDPPRMEPAATDPLTPPAPPAEPKPETPVAELPKPTPQLLSRHDELKERARQLLEATKREAREKERIRRQNSKEDKELNGVKNGDLSPKKIMTEEERQVMLRERARKLIADAKAGLVSPSSPLSPSRPPRAGAVEIISKQSVLDELAAEDRQSEDAKSTRSNSWGSDEARRSPLQSFTALVDSLSPEDHAADEGKESGSYIQSELEALEREQAAIDLKAAALEKQLRHVMEAADSTEEEDKLMSQWFNLVNKKNALLRRQMQLNILEQEEDLSRRCELLARELRLSLGVDEWRKTPGQKRRERLLLQELLSAVNERDRLVQEMDEQEKAIADDDAIERNLSHVEIQRKNNCILQ
Summary
Uniprot
H9JF73
A0A2W1B5C6
A0A2A4K8K7
A0A194Q6Q6
A0A2H1VQ82
A0A151WM95
+ More
F4WP88 A0A026WP25 A0A0L7QN88 A0A088A677 A0A3L8DWS0 A0A2A3E2Z2 E2B3V6 A0A0C9R2M6 A0A0P4VWA2 A0A224XAX8 E0VFP7 A0A0V0G592 A0A069DXH3 A0A1Y1NGF6 A0A1Y1NCC5 A0A1Y1NB72 A0A1W4WJY3 A0A232FKL6 A0A0A9WH21 A0A0N0BHC9 A0A1L8E5S0 D6WFM6 A0A1L8E5L0 A0A1B6E8M3 K7IZC5 A0A154PF74 A0A1B6DQ94 A0A1B0GHF6 V9I9S5 A0A1B6L0G6 U5EXG5 B4KML6 A0A0Q9XBH0 A0A182FTB0 B4JVN5 A0A0P5N828 A0A0Q9XMG3 A0A0P4YKC5 A0A0P6FFL4 A0A0P5LHT2 A0A0P6A7Z4 A0A0P5V0C6 A0A0P6DL35 A0A0P5RJR4 A0A0N8BQ44 A0A0N8D7K8 A0A0P5YCI8 A0A0P5IEZ8 A0A1J1J6R4
F4WP88 A0A026WP25 A0A0L7QN88 A0A088A677 A0A3L8DWS0 A0A2A3E2Z2 E2B3V6 A0A0C9R2M6 A0A0P4VWA2 A0A224XAX8 E0VFP7 A0A0V0G592 A0A069DXH3 A0A1Y1NGF6 A0A1Y1NCC5 A0A1Y1NB72 A0A1W4WJY3 A0A232FKL6 A0A0A9WH21 A0A0N0BHC9 A0A1L8E5S0 D6WFM6 A0A1L8E5L0 A0A1B6E8M3 K7IZC5 A0A154PF74 A0A1B6DQ94 A0A1B0GHF6 V9I9S5 A0A1B6L0G6 U5EXG5 B4KML6 A0A0Q9XBH0 A0A182FTB0 B4JVN5 A0A0P5N828 A0A0Q9XMG3 A0A0P4YKC5 A0A0P6FFL4 A0A0P5LHT2 A0A0P6A7Z4 A0A0P5V0C6 A0A0P6DL35 A0A0P5RJR4 A0A0N8BQ44 A0A0N8D7K8 A0A0P5YCI8 A0A0P5IEZ8 A0A1J1J6R4
Pubmed
EMBL
BABH01026837
BABH01026838
KZ150393
PZC71038.1
NWSH01000055
PCG80143.1
+ More
KQ459386 KPJ01227.1 ODYU01003768 SOQ42946.1 KQ982944 KYQ49012.1 GL888243 EGI64069.1 KK107139 EZA57698.1 KQ414856 KOC60095.1 QOIP01000003 RLU24887.1 KZ288430 PBC25844.1 GL445407 EFN89625.1 GBYB01002219 GBYB01002220 JAG71986.1 JAG71987.1 GDKW01000208 JAI56387.1 GFTR01008332 JAW08094.1 DS235123 EEB12203.1 GECL01002907 JAP03217.1 GBGD01000363 JAC88526.1 GEZM01007454 JAV95186.1 GEZM01007458 GEZM01007457 GEZM01007455 GEZM01007453 GEZM01007452 JAV95188.1 GEZM01007456 JAV95184.1 NNAY01000112 OXU30867.1 GBHO01036913 GBRD01001153 JAG06691.1 JAG64668.1 KQ435757 KOX75872.1 GFDF01000090 JAV13994.1 KQ971319 EFA00250.2 GFDF01000091 JAV13993.1 GEDC01003094 JAS34204.1 KQ434879 KZC09958.1 GEDC01009460 JAS27838.1 AJWK01004552 JR037568 JR037570 AEY57848.1 AEY57850.1 GEBQ01022832 JAT17145.1 GANO01002547 JAB57324.1 CH933808 EDW10863.1 KRG05541.1 CH916375 EDV98503.1 GDIQ01153209 JAK98516.1 KRG05543.1 GDIP01227563 JAI95838.1 GDIQ01049514 JAN45223.1 GDIQ01180296 JAK71429.1 GDIP01167221 GDIP01165670 GDIP01046043 JAM57672.1 GDIP01107426 JAL96288.1 GDIQ01076088 JAN18649.1 GDIQ01106025 JAL45701.1 GDIQ01155907 JAK95818.1 GDIP01060983 JAM42732.1 GDIP01060984 JAM42731.1 GDIQ01215593 JAK36132.1 CVRI01000074 CRL08109.1
KQ459386 KPJ01227.1 ODYU01003768 SOQ42946.1 KQ982944 KYQ49012.1 GL888243 EGI64069.1 KK107139 EZA57698.1 KQ414856 KOC60095.1 QOIP01000003 RLU24887.1 KZ288430 PBC25844.1 GL445407 EFN89625.1 GBYB01002219 GBYB01002220 JAG71986.1 JAG71987.1 GDKW01000208 JAI56387.1 GFTR01008332 JAW08094.1 DS235123 EEB12203.1 GECL01002907 JAP03217.1 GBGD01000363 JAC88526.1 GEZM01007454 JAV95186.1 GEZM01007458 GEZM01007457 GEZM01007455 GEZM01007453 GEZM01007452 JAV95188.1 GEZM01007456 JAV95184.1 NNAY01000112 OXU30867.1 GBHO01036913 GBRD01001153 JAG06691.1 JAG64668.1 KQ435757 KOX75872.1 GFDF01000090 JAV13994.1 KQ971319 EFA00250.2 GFDF01000091 JAV13993.1 GEDC01003094 JAS34204.1 KQ434879 KZC09958.1 GEDC01009460 JAS27838.1 AJWK01004552 JR037568 JR037570 AEY57848.1 AEY57850.1 GEBQ01022832 JAT17145.1 GANO01002547 JAB57324.1 CH933808 EDW10863.1 KRG05541.1 CH916375 EDV98503.1 GDIQ01153209 JAK98516.1 KRG05543.1 GDIP01227563 JAI95838.1 GDIQ01049514 JAN45223.1 GDIQ01180296 JAK71429.1 GDIP01167221 GDIP01165670 GDIP01046043 JAM57672.1 GDIP01107426 JAL96288.1 GDIQ01076088 JAN18649.1 GDIQ01106025 JAL45701.1 GDIQ01155907 JAK95818.1 GDIP01060983 JAM42732.1 GDIP01060984 JAM42731.1 GDIQ01215593 JAK36132.1 CVRI01000074 CRL08109.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF47576
SSF47576
Gene 3D
CDD
ProteinModelPortal
H9JF73
A0A2W1B5C6
A0A2A4K8K7
A0A194Q6Q6
A0A2H1VQ82
A0A151WM95
+ More
F4WP88 A0A026WP25 A0A0L7QN88 A0A088A677 A0A3L8DWS0 A0A2A3E2Z2 E2B3V6 A0A0C9R2M6 A0A0P4VWA2 A0A224XAX8 E0VFP7 A0A0V0G592 A0A069DXH3 A0A1Y1NGF6 A0A1Y1NCC5 A0A1Y1NB72 A0A1W4WJY3 A0A232FKL6 A0A0A9WH21 A0A0N0BHC9 A0A1L8E5S0 D6WFM6 A0A1L8E5L0 A0A1B6E8M3 K7IZC5 A0A154PF74 A0A1B6DQ94 A0A1B0GHF6 V9I9S5 A0A1B6L0G6 U5EXG5 B4KML6 A0A0Q9XBH0 A0A182FTB0 B4JVN5 A0A0P5N828 A0A0Q9XMG3 A0A0P4YKC5 A0A0P6FFL4 A0A0P5LHT2 A0A0P6A7Z4 A0A0P5V0C6 A0A0P6DL35 A0A0P5RJR4 A0A0N8BQ44 A0A0N8D7K8 A0A0P5YCI8 A0A0P5IEZ8 A0A1J1J6R4
F4WP88 A0A026WP25 A0A0L7QN88 A0A088A677 A0A3L8DWS0 A0A2A3E2Z2 E2B3V6 A0A0C9R2M6 A0A0P4VWA2 A0A224XAX8 E0VFP7 A0A0V0G592 A0A069DXH3 A0A1Y1NGF6 A0A1Y1NCC5 A0A1Y1NB72 A0A1W4WJY3 A0A232FKL6 A0A0A9WH21 A0A0N0BHC9 A0A1L8E5S0 D6WFM6 A0A1L8E5L0 A0A1B6E8M3 K7IZC5 A0A154PF74 A0A1B6DQ94 A0A1B0GHF6 V9I9S5 A0A1B6L0G6 U5EXG5 B4KML6 A0A0Q9XBH0 A0A182FTB0 B4JVN5 A0A0P5N828 A0A0Q9XMG3 A0A0P4YKC5 A0A0P6FFL4 A0A0P5LHT2 A0A0P6A7Z4 A0A0P5V0C6 A0A0P6DL35 A0A0P5RJR4 A0A0N8BQ44 A0A0N8D7K8 A0A0P5YCI8 A0A0P5IEZ8 A0A1J1J6R4
PDB
2D89
E-value=1.27103e-25,
Score=293
Ontologies
GO
PANTHER
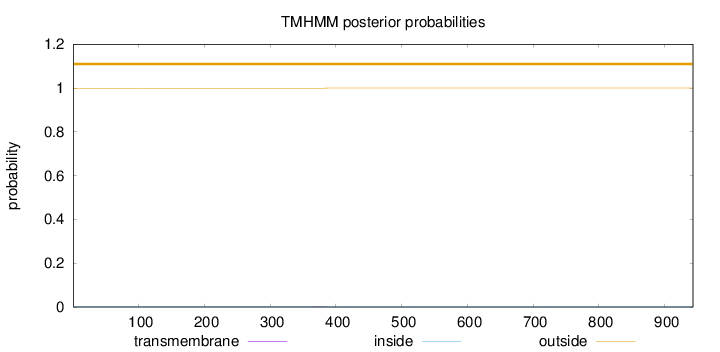
Topology
Length:
943
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01363
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00071
outside
1 - 943
Population Genetic Test Statistics
Pi
202.380587
Theta
20.54344
Tajima's D
1.328947
CLR
0.194765
CSRT
0.750512474376281
Interpretation
Uncertain
