Pre Gene Modal
BGIBMGA008154
Annotation
PREDICTED:_dynamin-like_120_kDa_protein?_mitochondrial_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.15
Sequence
CDS
ATGTCCAGAATACTACATAATCGTGTAAGATCATCATTTAAGAACACATCATGGGTCAGCATCGACAAACGAGCAGTAGCATTCTCAATATGGACTGGACGTAGCATGCTACTGTACAAACCAATAGATACGAGCCATGTGCAGAGAAGATGCTATGGTATGCTTGTTGCAAGGGCTATTAGAGGAGTGTTGAAAATTCGTTACTTAGTTCTCGGTGGTGCCGTTGGTGGAGGCATGACGTTGAATAAGAAATATGCCGAGTGGAAGGATGGTCTTCCAGATATGGGCTGGTTAAATGACCTGCTCCCCGACAATGAACAATGGGATAGATTTACAACTACACTGATTACAGCAAAAGATAAAATCGGTGATCAATTGCAGATTGATCCACGACTCCGTGAAGCTGGTGTTGCCAGGACCACCGAGCTGCGCGAGTGGTTAGCTCAGAGGTACGAAGATGCCGTGGCGGCCGCAGCCGTTAACAATCAACAACGATTAGTGTATTGTTCAGAGCCGGCTCCGAAGATAGTGAACAACCTCCAGAGCAGGCCCGCGTCGCCGCCGCCCGCGCGCTCCGCCGACGACGCCGCGCACTACGGTGAGTCCGCGACTTGCTCACATTTCTATGTCACTCAGTTTACCCGGGTGTATCCGTAG
Protein
MSRILHNRVRSSFKNTSWVSIDKRAVAFSIWTGRSMLLYKPIDTSHVQRRCYGMLVARAIRGVLKIRYLVLGGAVGGGMTLNKKYAEWKDGLPDMGWLNDLLPDNEQWDRFTTTLITAKDKIGDQLQIDPRLREAGVARTTELREWLAQRYEDAVAAAAVNNQQRLVYCSEPAPKIVNNLQSRPASPPPARSADDAAHYGESATCSHFYVTQFTRVYP
Summary
Similarity
Belongs to the TRAFAC class dynamin-like GTPase superfamily. Dynamin/Fzo/YdjA family.
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
Ontologies
PANTHER
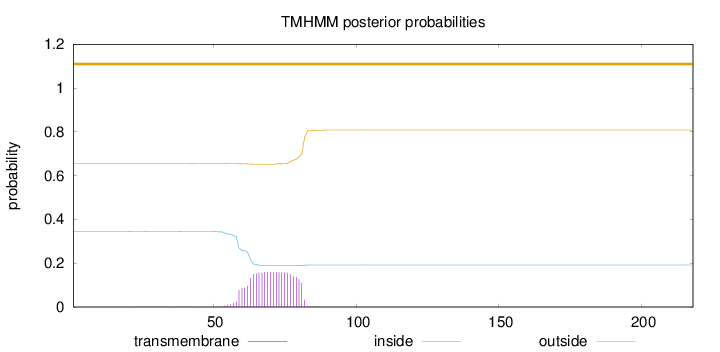
Topology
Length:
218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.31837
Exp number, first 60 AAs:
0.24766
Total prob of N-in:
0.34438
outside
1 - 218
Population Genetic Test Statistics
Pi
26.86408
Theta
22.423575
Tajima's D
0.556495
CLR
0.640893
CSRT
0.535923203839808
Interpretation
Uncertain
