Pre Gene Modal
BGIBMGA008154
Annotation
PREDICTED:_dynamin-like_120_kDa_protein?_mitochondrial_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.341
Sequence
CDS
ATGATGAAGAAACGTGTCCACGCCTTGCAAGAGGAAGTACTGGCTCTACAGGCGCGCTACCAGAGGGAGTTGCAGCGGCTAGAACAAGAGAACAGAGAGCTGAGGCAGCAGAACATGCTGCTGAAACAAGGCAGGCAACCTTCCACTAAGAAGATGAAGCGGTCACTGATCGACATGTACTCGTCCGTGCTGGACGAGCTGGCGGGCTGGGAGGCGGACGGCGCGGACCGGCTGCCGCGGGTGGTGGTGGTGGGCGACCAGTCCGCCGGCAAGACCTCCGTGCTGGAGATGATCGCGCAGGCCAGGATCTTCCCGCGCGGCGCTGGCGAGATGTGCACCAGAGCCCCGGTGAAGGTGACGCTGTCCGAGGGCCCGTACCACGTAGCGCAGTTCAGGGACTCCTCCAGAGAGTTTGATCTCAATAAGGAATCAGATTTAGCTGACTTAAGGAAGGAGGTGGAGCTGCGCATGCGCAACAGCGTGCGCGGCGGCCGCACGGTCTCGTCGGACGTCATCTCGATGTCGGTGAAGGGGCCCGGCCTGCACCGGATGGTGCTGGTGGACCTGCCCGGAGTCATCTCGACTCAAACCGTGGACATGGCGGCGGACACTCGCGAGGCCATCAAGCAGATGACCAGACAGTACATGGACAACCCCAACGCCATCATCCTGTGCATCCAGGACGGGTCAGTGGACGCAGAGCGCAGCAACGTGACCGACCTGGTGTCCTCCTGTGACCCACAGGGGAAACGAACGATCTTCGTGCTCACCAAGGTGGATCTGGCCGAAGAGAACCTCGCCAATCCGAACCGGATCCGCCGCATCCTGGAGGGCAAGCTGTTCCCGATGAAGGCGCTGGGCTACTACGCCGTGGTGACGGGCCGCTCGCGCCGGGACGACTCCATCCAGGGCATCAGGGAGTACGAGGAGAGATTCTTCAGCTCCTCGAAACTATTCAAGAATGGTCTGGTGACCCCATCGCAGGTGACCACCAGGAACCTGTCGCTGGCCGTCGCCGAGTGCTTCTGGAGGATGGTGCGCGCGACCGTCGAGCAGCAAGCCGACGCCTTCAAAGCCATGAGGTTCAACCTGGAGACCGAATGGAAGAACACGTTCCCGAGGCAGCGCGAGCTGGACCGCGACGAGCTGTTCGAGCGCGCGCGCGCCGAGCTGCTGGACCAGGCCGCCGAGCTGTCGGCCGTGCCCGCCCAGCGCTGGGAGCACGCGCTGCGGGACGCGCTGTGGGACCACGCCGCGCCCACCGTCATGGACCGCATCTACCTGCCCGCCGCCGCCGCCGCGCACGACGATCAAGACAAGTTTAACACGATAGTGGACATAAAGCTGCGCGAGTGGGCGGAGCGCGAGCTGCCCGGCACGTCGGTGCAGGCCGGGCGGGAGGCCCTGCGCGCCGAGTTCTCGCGCCTCACGGAGCCCGCGGAGCCCGAGCCGCTGTTCCGCGGGCTGCGCGAGCACGCCGTGCGCGAGGCGCTGCTGCGGCACGACTGGGAGGAGCGCGCGCAGGACGTGCTGCGCGTGCTGCAGCTGAACGCGCTGCAGGACCGCTGCGTGAGCGCGCGCGCGCACTGGGACCGCGCCGTGGCGCTGCTGGAGGCCGCGCTGGGGGACCGCCTGGGGGCCGCGCGGGACCGTCTGCGGGACCTCACCGGTCCGGGATGGCGGGAGCGCTGGCTGTCTTGGAGCAGCCCCACGGAGGAGCAGCGGCGCCGCGCCCTGCTGGCCGCCGAGCTGGAGCAGCTGGGCACCGCGCGCCCGCAGCTCACGTACGACGAGGTGGTGGCCGTGCGGGACAACCTGCGCCGGCAGGGCGTGGAGGTGGACAACGACTACATCAGGGAGACGTGGAAGCCGCTGTACCTGAGGAAGTTCTTGGAGCAGGCACTGACCCGCGCCCGGGACTGCAGGAAGAGCTTCTACTTGTACAGCCAACAGAACGGCGCCGGGAAGGAGTGTTGCGAGGTGGTGCTGTTCTGGCGCATCCAGCAGACCCTGGAGACCACGGCCAACGCTCTGCGACAGCAGATCGGGAACCGGGAGGCCGCCAGGCTCGACAGGGAGCTGCGGGAAGTGCTCGATGAGATGGCGGCCGACGCGGACCTCAAGAAGCAGCTGCTGTCCGGCAGGCGGGTGGAGCTCGCAGAGGAACTAAAACGAGTGCGGAAAGTACAAGAGAAATTAGAGGAATTTATCGAAGCACTTAACAAAGAAAAGTAG
Protein
MMKKRVHALQEEVLALQARYQRELQRLEQENRELRQQNMLLKQGRQPSTKKMKRSLIDMYSSVLDELAGWEADGADRLPRVVVVGDQSAGKTSVLEMIAQARIFPRGAGEMCTRAPVKVTLSEGPYHVAQFRDSSREFDLNKESDLADLRKEVELRMRNSVRGGRTVSSDVISMSVKGPGLHRMVLVDLPGVISTQTVDMAADTREAIKQMTRQYMDNPNAIILCIQDGSVDAERSNVTDLVSSCDPQGKRTIFVLTKVDLAEENLANPNRIRRILEGKLFPMKALGYYAVVTGRSRRDDSIQGIREYEERFFSSSKLFKNGLVTPSQVTTRNLSLAVAECFWRMVRATVEQQADAFKAMRFNLETEWKNTFPRQRELDRDELFERARAELLDQAAELSAVPAQRWEHALRDALWDHAAPTVMDRIYLPAAAAAHDDQDKFNTIVDIKLREWAERELPGTSVQAGREALRAEFSRLTEPAEPEPLFRGLREHAVREALLRHDWEERAQDVLRVLQLNALQDRCVSARAHWDRAVALLEAALGDRLGAARDRLRDLTGPGWRERWLSWSSPTEEQRRRALLAAELEQLGTARPQLTYDEVVAVRDNLRRQGVEVDNDYIRETWKPLYLRKFLEQALTRARDCRKSFYLYSQQNGAGKECCEVVLFWRIQQTLETTANALRQQIGNREAARLDRELREVLDEMAADADLKKQLLSGRRVELAEELKRVRKVQEKLEEFIEALNKEK
Summary
Similarity
Belongs to the TRAFAC class dynamin-like GTPase superfamily. Dynamin/Fzo/YdjA family.
Uniprot
A0A2W1B809
A0A194Q768
A0A1Y1L9W0
A0A1Y1LFR8
A0A1Y1LEL3
K7J9S6
+ More
A0A2A3ED40 A0A088AMM2 D6WKP0 A0A2J7QGD8 K7J9S7 K7J9S8 A0A139WI82 A0A232F5P9 A0A3L8D5N3 A0A026W2E8 A0A2J7QG87 A0A2J7QG90 A0A2J7QG95 A0A2J7QGA7 F4X7H0 E2AJU3 A0A151JWS6 A0A158NN27 A0A158NN26 A0A158NN28 A0A182HCJ9 A0A182HCB7 A0A1S4F5T3 Q17EC7 A0A2M4AG27 A0A0C9QWU4 A0A182QIA0 A0A2M4AEA2 A0A182FLL7 A0A3F2YWU8 A0A2M4AE80 A0A084VD28 Q7QHP4 A0A2M4ADD6 A0A2M4ADB6 W5JSG8 U5EJB3 E9IDV8 A0A182Y6V7 A0A2M4BC90 A0A182UHC0 A0A2M4BC49 A0A182WYT1 A0A2M3Z351 A0A2M3ZHC5 A0A182HYA3 A0A182MMK4 A0A182RL05 A0A182UY49 A0A182JQZ4 A0A0C9QXH8 A0A182LHG1 A0A0L7RAM3 A0A182NT16 A0A0N0BFI9 A0A182J678 A0A310SLL7 A0A2H1WG64 A0A1Y1LBZ5 A0A1Q3G3D9 A0A1Q3G377 E2BGA5 A0A1Q3FXV5 A0A336M3C7 A0A336MA42 A0A0N7Z9Y2 A0A0P4VUU6 A0A0J7L7N7 A0A0L0CBP8 A0A1B0CGP7 A0A1L8E076 A0A1L8DZY5 A0A1L8E097 A0A1L8E013 A0A195C471 E0VXD9 A0A0A1WQ08 A0A0A1X945 A0A0K8VGV8 W8BRN5 W8C920 A0A0K8VXW8 A0A034W9G1 A0A034W7T6 A0A0K8U291 A0A164Y781 A0A0Q9X887 A0A0K8V3Y8 A0A0P6GMI8 T1IBX0 A0A1I8PFT8 A0A0P5ZTU7 E9FYT2 A0A151IV15
A0A2A3ED40 A0A088AMM2 D6WKP0 A0A2J7QGD8 K7J9S7 K7J9S8 A0A139WI82 A0A232F5P9 A0A3L8D5N3 A0A026W2E8 A0A2J7QG87 A0A2J7QG90 A0A2J7QG95 A0A2J7QGA7 F4X7H0 E2AJU3 A0A151JWS6 A0A158NN27 A0A158NN26 A0A158NN28 A0A182HCJ9 A0A182HCB7 A0A1S4F5T3 Q17EC7 A0A2M4AG27 A0A0C9QWU4 A0A182QIA0 A0A2M4AEA2 A0A182FLL7 A0A3F2YWU8 A0A2M4AE80 A0A084VD28 Q7QHP4 A0A2M4ADD6 A0A2M4ADB6 W5JSG8 U5EJB3 E9IDV8 A0A182Y6V7 A0A2M4BC90 A0A182UHC0 A0A2M4BC49 A0A182WYT1 A0A2M3Z351 A0A2M3ZHC5 A0A182HYA3 A0A182MMK4 A0A182RL05 A0A182UY49 A0A182JQZ4 A0A0C9QXH8 A0A182LHG1 A0A0L7RAM3 A0A182NT16 A0A0N0BFI9 A0A182J678 A0A310SLL7 A0A2H1WG64 A0A1Y1LBZ5 A0A1Q3G3D9 A0A1Q3G377 E2BGA5 A0A1Q3FXV5 A0A336M3C7 A0A336MA42 A0A0N7Z9Y2 A0A0P4VUU6 A0A0J7L7N7 A0A0L0CBP8 A0A1B0CGP7 A0A1L8E076 A0A1L8DZY5 A0A1L8E097 A0A1L8E013 A0A195C471 E0VXD9 A0A0A1WQ08 A0A0A1X945 A0A0K8VGV8 W8BRN5 W8C920 A0A0K8VXW8 A0A034W9G1 A0A034W7T6 A0A0K8U291 A0A164Y781 A0A0Q9X887 A0A0K8V3Y8 A0A0P6GMI8 T1IBX0 A0A1I8PFT8 A0A0P5ZTU7 E9FYT2 A0A151IV15
Pubmed
EMBL
KZ150293
PZC71568.1
KQ459386
KPJ01229.1
GEZM01061523
JAV70414.1
+ More
GEZM01061521 JAV70416.1 GEZM01061522 JAV70415.1 AAZX01005171 KZ288280 PBC29665.1 KQ971342 EFA02998.2 NEVH01014835 PNF27603.1 KYB27693.1 NNAY01000869 OXU26134.1 QOIP01000013 RLU15539.1 KK107467 EZA50265.1 PNF27607.1 PNF27606.1 PNF27604.1 PNF27608.1 GL888840 EGI57608.1 GL440100 EFN66259.1 KQ981645 KYN38678.1 ADTU01020949 JXUM01127563 KQ567205 KXJ69556.1 JXUM01127004 KQ567136 KXJ69605.1 CH477283 EAT44850.1 GGFK01006424 MBW39745.1 GBYB01000170 JAG69937.1 AXCN02001792 GGFK01005795 MBW39116.1 GGFK01005773 MBW39094.1 ATLV01011135 KE524643 KFB35872.1 AAAB01008816 EAA05165.3 GGFK01005468 MBW38789.1 GGFK01005464 MBW38785.1 ADMH02000297 ETN67071.1 GANO01002295 JAB57576.1 GL762535 EFZ21270.1 GGFJ01001496 MBW50637.1 GGFJ01001451 MBW50592.1 GGFM01002196 MBW22947.1 GGFM01007172 MBW27923.1 APCN01004510 AXCM01005991 GBYB01005372 JAG75139.1 KQ414618 KOC67899.1 KQ435798 KOX73411.1 KQ761208 OAD57986.1 ODYU01008117 SOQ51444.1 GEZM01061520 JAV70418.1 GFDL01000766 JAV34279.1 GFDL01000795 JAV34250.1 GL448138 EFN85268.1 GFDL01002654 JAV32391.1 UFQT01000292 SSX22907.1 SSX22908.1 GDRN01108071 JAI57320.1 GDRN01108074 GDRN01108073 JAI57317.1 LBMM01000366 KMQ99191.1 JRES01000644 KNC29665.1 AJWK01011531 GFDF01002017 JAV12067.1 GFDF01002016 JAV12068.1 GFDF01002000 JAV12084.1 GFDF01002001 JAV12083.1 KQ978344 KYM94968.1 DS235830 EEB18045.1 GBXI01013561 JAD00731.1 GBXI01006866 JAD07426.1 GDHF01029054 GDHF01014217 JAI23260.1 JAI38097.1 GAMC01007022 JAB99533.1 GAMC01007021 JAB99534.1 GDHF01009830 GDHF01008884 JAI42484.1 JAI43430.1 GAKP01008192 JAC50760.1 GAKP01008193 JAC50759.1 GDHF01031661 JAI20653.1 LRGB01000930 KZS14950.1 CH933808 KRG04598.1 GDHF01019039 JAI33275.1 GDIQ01033157 JAN61580.1 ACPB03000857 GDIP01039696 JAM64019.1 GL732527 EFX87719.1 KQ980928 KYN11370.1
GEZM01061521 JAV70416.1 GEZM01061522 JAV70415.1 AAZX01005171 KZ288280 PBC29665.1 KQ971342 EFA02998.2 NEVH01014835 PNF27603.1 KYB27693.1 NNAY01000869 OXU26134.1 QOIP01000013 RLU15539.1 KK107467 EZA50265.1 PNF27607.1 PNF27606.1 PNF27604.1 PNF27608.1 GL888840 EGI57608.1 GL440100 EFN66259.1 KQ981645 KYN38678.1 ADTU01020949 JXUM01127563 KQ567205 KXJ69556.1 JXUM01127004 KQ567136 KXJ69605.1 CH477283 EAT44850.1 GGFK01006424 MBW39745.1 GBYB01000170 JAG69937.1 AXCN02001792 GGFK01005795 MBW39116.1 GGFK01005773 MBW39094.1 ATLV01011135 KE524643 KFB35872.1 AAAB01008816 EAA05165.3 GGFK01005468 MBW38789.1 GGFK01005464 MBW38785.1 ADMH02000297 ETN67071.1 GANO01002295 JAB57576.1 GL762535 EFZ21270.1 GGFJ01001496 MBW50637.1 GGFJ01001451 MBW50592.1 GGFM01002196 MBW22947.1 GGFM01007172 MBW27923.1 APCN01004510 AXCM01005991 GBYB01005372 JAG75139.1 KQ414618 KOC67899.1 KQ435798 KOX73411.1 KQ761208 OAD57986.1 ODYU01008117 SOQ51444.1 GEZM01061520 JAV70418.1 GFDL01000766 JAV34279.1 GFDL01000795 JAV34250.1 GL448138 EFN85268.1 GFDL01002654 JAV32391.1 UFQT01000292 SSX22907.1 SSX22908.1 GDRN01108071 JAI57320.1 GDRN01108074 GDRN01108073 JAI57317.1 LBMM01000366 KMQ99191.1 JRES01000644 KNC29665.1 AJWK01011531 GFDF01002017 JAV12067.1 GFDF01002016 JAV12068.1 GFDF01002000 JAV12084.1 GFDF01002001 JAV12083.1 KQ978344 KYM94968.1 DS235830 EEB18045.1 GBXI01013561 JAD00731.1 GBXI01006866 JAD07426.1 GDHF01029054 GDHF01014217 JAI23260.1 JAI38097.1 GAMC01007022 JAB99533.1 GAMC01007021 JAB99534.1 GDHF01009830 GDHF01008884 JAI42484.1 JAI43430.1 GAKP01008192 JAC50760.1 GAKP01008193 JAC50759.1 GDHF01031661 JAI20653.1 LRGB01000930 KZS14950.1 CH933808 KRG04598.1 GDHF01019039 JAI33275.1 GDIQ01033157 JAN61580.1 ACPB03000857 GDIP01039696 JAM64019.1 GL732527 EFX87719.1 KQ980928 KYN11370.1
Proteomes
UP000053268
UP000002358
UP000242457
UP000005203
UP000007266
UP000235965
+ More
UP000215335 UP000279307 UP000053097 UP000007755 UP000000311 UP000078541 UP000005205 UP000069940 UP000249989 UP000008820 UP000075886 UP000069272 UP000030765 UP000007062 UP000000673 UP000076408 UP000075902 UP000076407 UP000075840 UP000075883 UP000075900 UP000075903 UP000075881 UP000075882 UP000053825 UP000075884 UP000053105 UP000075880 UP000008237 UP000036403 UP000037069 UP000092461 UP000078542 UP000009046 UP000076858 UP000009192 UP000015103 UP000095300 UP000000305 UP000078492
UP000215335 UP000279307 UP000053097 UP000007755 UP000000311 UP000078541 UP000005205 UP000069940 UP000249989 UP000008820 UP000075886 UP000069272 UP000030765 UP000007062 UP000000673 UP000076408 UP000075902 UP000076407 UP000075840 UP000075883 UP000075900 UP000075903 UP000075881 UP000075882 UP000053825 UP000075884 UP000053105 UP000075880 UP000008237 UP000036403 UP000037069 UP000092461 UP000078542 UP000009046 UP000076858 UP000009192 UP000015103 UP000095300 UP000000305 UP000078492
PRIDE
Pfam
PF00350 Dynamin_N
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A2W1B809
A0A194Q768
A0A1Y1L9W0
A0A1Y1LFR8
A0A1Y1LEL3
K7J9S6
+ More
A0A2A3ED40 A0A088AMM2 D6WKP0 A0A2J7QGD8 K7J9S7 K7J9S8 A0A139WI82 A0A232F5P9 A0A3L8D5N3 A0A026W2E8 A0A2J7QG87 A0A2J7QG90 A0A2J7QG95 A0A2J7QGA7 F4X7H0 E2AJU3 A0A151JWS6 A0A158NN27 A0A158NN26 A0A158NN28 A0A182HCJ9 A0A182HCB7 A0A1S4F5T3 Q17EC7 A0A2M4AG27 A0A0C9QWU4 A0A182QIA0 A0A2M4AEA2 A0A182FLL7 A0A3F2YWU8 A0A2M4AE80 A0A084VD28 Q7QHP4 A0A2M4ADD6 A0A2M4ADB6 W5JSG8 U5EJB3 E9IDV8 A0A182Y6V7 A0A2M4BC90 A0A182UHC0 A0A2M4BC49 A0A182WYT1 A0A2M3Z351 A0A2M3ZHC5 A0A182HYA3 A0A182MMK4 A0A182RL05 A0A182UY49 A0A182JQZ4 A0A0C9QXH8 A0A182LHG1 A0A0L7RAM3 A0A182NT16 A0A0N0BFI9 A0A182J678 A0A310SLL7 A0A2H1WG64 A0A1Y1LBZ5 A0A1Q3G3D9 A0A1Q3G377 E2BGA5 A0A1Q3FXV5 A0A336M3C7 A0A336MA42 A0A0N7Z9Y2 A0A0P4VUU6 A0A0J7L7N7 A0A0L0CBP8 A0A1B0CGP7 A0A1L8E076 A0A1L8DZY5 A0A1L8E097 A0A1L8E013 A0A195C471 E0VXD9 A0A0A1WQ08 A0A0A1X945 A0A0K8VGV8 W8BRN5 W8C920 A0A0K8VXW8 A0A034W9G1 A0A034W7T6 A0A0K8U291 A0A164Y781 A0A0Q9X887 A0A0K8V3Y8 A0A0P6GMI8 T1IBX0 A0A1I8PFT8 A0A0P5ZTU7 E9FYT2 A0A151IV15
A0A2A3ED40 A0A088AMM2 D6WKP0 A0A2J7QGD8 K7J9S7 K7J9S8 A0A139WI82 A0A232F5P9 A0A3L8D5N3 A0A026W2E8 A0A2J7QG87 A0A2J7QG90 A0A2J7QG95 A0A2J7QGA7 F4X7H0 E2AJU3 A0A151JWS6 A0A158NN27 A0A158NN26 A0A158NN28 A0A182HCJ9 A0A182HCB7 A0A1S4F5T3 Q17EC7 A0A2M4AG27 A0A0C9QWU4 A0A182QIA0 A0A2M4AEA2 A0A182FLL7 A0A3F2YWU8 A0A2M4AE80 A0A084VD28 Q7QHP4 A0A2M4ADD6 A0A2M4ADB6 W5JSG8 U5EJB3 E9IDV8 A0A182Y6V7 A0A2M4BC90 A0A182UHC0 A0A2M4BC49 A0A182WYT1 A0A2M3Z351 A0A2M3ZHC5 A0A182HYA3 A0A182MMK4 A0A182RL05 A0A182UY49 A0A182JQZ4 A0A0C9QXH8 A0A182LHG1 A0A0L7RAM3 A0A182NT16 A0A0N0BFI9 A0A182J678 A0A310SLL7 A0A2H1WG64 A0A1Y1LBZ5 A0A1Q3G3D9 A0A1Q3G377 E2BGA5 A0A1Q3FXV5 A0A336M3C7 A0A336MA42 A0A0N7Z9Y2 A0A0P4VUU6 A0A0J7L7N7 A0A0L0CBP8 A0A1B0CGP7 A0A1L8E076 A0A1L8DZY5 A0A1L8E097 A0A1L8E013 A0A195C471 E0VXD9 A0A0A1WQ08 A0A0A1X945 A0A0K8VGV8 W8BRN5 W8C920 A0A0K8VXW8 A0A034W9G1 A0A034W7T6 A0A0K8U291 A0A164Y781 A0A0Q9X887 A0A0K8V3Y8 A0A0P6GMI8 T1IBX0 A0A1I8PFT8 A0A0P5ZTU7 E9FYT2 A0A151IV15
PDB
6DI7
E-value=9.98283e-27,
Score=301
Ontologies
GO
PANTHER
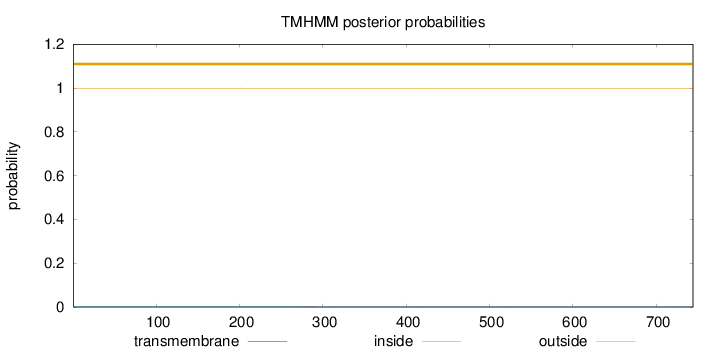
Topology
Length:
744
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0043
Exp number, first 60 AAs:
6e-05
Total prob of N-in:
0.00066
outside
1 - 744
Population Genetic Test Statistics
Pi
360.337623
Theta
211.316776
Tajima's D
2.519566
CLR
0.062996
CSRT
0.946902654867257
Interpretation
Uncertain
