Gene
KWMTBOMO14271
Pre Gene Modal
BGIBMGA008155
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_rho_guanine_nucleotide_exchange_factor_11_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.813
Sequence
CDS
ATGGAGGAGGATGACTCCCCCGCCTCGGACACGTCCACATACTCCGGGCTGTTCTCGAACCTGCGGTCGGTGCGCGAGTCCAAGGCCAGGACCGCGGTGCTGCTAAACTGGCTACTGGGAGAAAGGAGATGCGCGTGCGCCAGTCTGCTGCTGATCCTCACGGACGCGTACCGGTCGGCGAGTGGCGTGACGGCGGCCACGCTGCGCCGCTGGGCCTACGAGATACACTCCACGTTCCTCATGCCCAACGCGGTTCTACAGCTGAATGGCGTCGACGAAAACATGGCGAATGAAATCGAACATATCCTAGTTAACGAGCACGACAAGGAGGAGCTGGTCCGCAACGTGTTCCGCAAGGCGCGGCAGAAGGCGAAGGAGGAGCTGAGCTCGCAGCTGTCGGAGTTCCAGGTGAAGCGGCAGGTGGGGCTCGGCACGCTGTACGGCCCGCACGACGCGCTGCTGGAGCGCTGCGACCTGGACAAGGCGCAGGAGCTGGTGGTGGTGGAGCAGGTGATGACGTCACAGTTGCGCGCGGTGAGCGCCGCGTCCCCCGCGCCCGCCGCCCCCGCCCCCTCGCCCGCCGCCGCGCAGGACGCGCACAACACGCTGCTGGCCGCGCTCATCGCCGCCGCGCTCGCGCTGCACTGCCGGCCCGCGTCCGTGGCCGCCGCCGTGGGGGGCGCCCGCGCCTCCTTCCTGCAGAGGGACAAGCTGTACAAGCAGAGGGTCAAGCAGATCATGAAGCAGCATCCCCAGCCGTTCGTGGTGAGGGGCCACCACCTGCAGCTGCAGGCGCTGACGTCGGTGGGGCACTGCCACCGCTGCGACCACGTCATCTGGGGCCTGGCGCCGCAAGCCTACGTGTGTGCCGATTGCAAGCTGCGGGTCCACCGCGCCTGCGCCAAGTCCGTGGAGGAGGGCTGCTGCCTCGACGGGGAGCACCACACCAACAGGATATCACGGTTCATGGAGCGGATGCATCCCAACAATCAACCCAACGCTCAAGACGTGAACGACAAGAAATCAAGAAAGGCTTCGGGCACGGCGCATTTCCTCAACATGGAGCGCAGTTTCCGCAAGGCTGAAGACGAGGCACCCTGGGACGCGATCCTCGCACCAGCTCAAGGATAA
Protein
MEEDDSPASDTSTYSGLFSNLRSVRESKARTAVLLNWLLGERRCACASLLLILTDAYRSASGVTAATLRRWAYEIHSTFLMPNAVLQLNGVDENMANEIEHILVNEHDKEELVRNVFRKARQKAKEELSSQLSEFQVKRQVGLGTLYGPHDALLERCDLDKAQELVVVEQVMTSQLRAVSAASPAPAAPAPSPAAAQDAHNTLLAALIAAALALHCRPASVAAAVGGARASFLQRDKLYKQRVKQIMKQHPQPFVVRGHHLQLQALTSVGHCHRCDHVIWGLAPQAYVCADCKLRVHRACAKSVEEGCCLDGEHHTNRISRFMERMHPNNQPNAQDVNDKKSRKASGTAHFLNMERSFRKAEDEAPWDAILAPAQG
Summary
Uniprot
H9JF58
A0A1B6JC30
A0A1B6DCQ4
A0A1B6CCT4
A0A1B6CEY4
A0A1B6C0U4
+ More
A0A1B6LSD5 A0A1B6L3G5 A0A1B6L264 A0A1B6MR53 A0A1B6J8S6 A0A224X591 A0A1B6HFE4 A0A232EW19 K7JHC5 A0A0L7RH29 A0A310SED8 A0A154PFD2 E0VFR4 A0A0C9S1B8 A0A0C9RYX7 A0A336KLF3 A0A336MEI9 A0A0M8ZYZ7 A0A2H8TCQ7 A0A164RBB5 A0A1W4UZR7 A0A1W4VDC9 A0A146LHP2 A0A2M3YZF4 A0A1W4VC14 W5JW08 A0A2M4CZJ0 A0A146LHC1 A0A2M4A418 A0A2M4BA38 A0A0A9Y0I0 A0A0A9Y211 A0A2M4A2Y9 A0A0P5PQ09 A0A0P6FDE1 A0A0P6EL40 A0A0A9WME6 A0A2M4CLI9 A0A0P5PG33 A0A0A9Y833 A0A182FVX8 A0A0P5ZLI1 A0A2M4B8X3 A0A0P5CMD4 A0A0P5IH19 A0A0Q5VM08 A0A0N8E7G0 A0A0P5GD22 A0A0P6EHW8 B3NP30 A0A2S2NCX6 A0A0P5VRN3 A0A0P5VNJ4 A0A0P4Y9Z7 A0A0P6H2V5 A0A0N8DAE5 A0A0P5QUN2 A0A0P6CRF7 A0A0P5CTM7 A0A0N8EGX1 A0A0P5CW19 A0A0N8AAJ7 A0A0N8AHE6 A0A0N8EFF9 A0A0P4ZT61 A0A0P5BF15 A0A0P5PDU4 A0A0P6C0W5 A0A2M3ZZH5 A0A0P5D8V5 A0A0P5YXE7 A1ZAN6 A0A0P5YAW3 A0A0P5BYH5 A0A0P5BF40 A0A0P5YV04 Q8T9E3 A0A0P5Y2W1 A0A0P5ALS5 A0A0P5E7C4 A0A0P5UPY8 A0A0B4K7T7 O44381 A0A0P5WDP8
A0A1B6LSD5 A0A1B6L3G5 A0A1B6L264 A0A1B6MR53 A0A1B6J8S6 A0A224X591 A0A1B6HFE4 A0A232EW19 K7JHC5 A0A0L7RH29 A0A310SED8 A0A154PFD2 E0VFR4 A0A0C9S1B8 A0A0C9RYX7 A0A336KLF3 A0A336MEI9 A0A0M8ZYZ7 A0A2H8TCQ7 A0A164RBB5 A0A1W4UZR7 A0A1W4VDC9 A0A146LHP2 A0A2M3YZF4 A0A1W4VC14 W5JW08 A0A2M4CZJ0 A0A146LHC1 A0A2M4A418 A0A2M4BA38 A0A0A9Y0I0 A0A0A9Y211 A0A2M4A2Y9 A0A0P5PQ09 A0A0P6FDE1 A0A0P6EL40 A0A0A9WME6 A0A2M4CLI9 A0A0P5PG33 A0A0A9Y833 A0A182FVX8 A0A0P5ZLI1 A0A2M4B8X3 A0A0P5CMD4 A0A0P5IH19 A0A0Q5VM08 A0A0N8E7G0 A0A0P5GD22 A0A0P6EHW8 B3NP30 A0A2S2NCX6 A0A0P5VRN3 A0A0P5VNJ4 A0A0P4Y9Z7 A0A0P6H2V5 A0A0N8DAE5 A0A0P5QUN2 A0A0P6CRF7 A0A0P5CTM7 A0A0N8EGX1 A0A0P5CW19 A0A0N8AAJ7 A0A0N8AHE6 A0A0N8EFF9 A0A0P4ZT61 A0A0P5BF15 A0A0P5PDU4 A0A0P6C0W5 A0A2M3ZZH5 A0A0P5D8V5 A0A0P5YXE7 A1ZAN6 A0A0P5YAW3 A0A0P5BYH5 A0A0P5BF40 A0A0P5YV04 Q8T9E3 A0A0P5Y2W1 A0A0P5ALS5 A0A0P5E7C4 A0A0P5UPY8 A0A0B4K7T7 O44381 A0A0P5WDP8
Pubmed
EMBL
BABH01026813
BABH01026814
GECU01010962
JAS96744.1
GEDC01013923
JAS23375.1
+ More
GEDC01026077 JAS11221.1 GEDC01025301 JAS11997.1 GEDC01030180 JAS07118.1 GEBQ01013488 JAT26489.1 GEBQ01021782 JAT18195.1 GEBQ01022246 JAT17731.1 GEBQ01001563 JAT38414.1 GECU01012161 JAS95545.1 GFTR01008761 JAW07665.1 GECU01034344 JAS73362.1 NNAY01001923 OXU22552.1 KQ414591 KOC70282.1 KQ797310 OAD46959.1 KQ434879 KZC09940.1 DS235123 EEB12220.1 GBYB01014566 JAG84333.1 GBYB01014570 JAG84337.1 UFQS01000449 UFQT01000449 SSX04115.1 SSX24480.1 SSX04113.1 SSX24478.1 KQ435794 KOX73980.1 GFXV01000080 MBW11885.1 LRGB01002190 KZS08498.1 GDHC01012173 JAQ06456.1 GGFM01000908 MBW21659.1 ADMH02000286 ETN67114.1 GGFL01006566 MBW70744.1 GDHC01011085 JAQ07544.1 GGFK01002174 MBW35495.1 GGFJ01000710 MBW49851.1 GBHO01017925 JAG25679.1 GBHO01022022 GBHO01017923 JAG21582.1 JAG25681.1 GGFK01001808 MBW35129.1 GDIQ01131260 JAL20466.1 GDIQ01065438 JAN29299.1 GDIQ01062441 JAN32296.1 GBHO01034007 JAG09597.1 GGFL01001957 MBW66135.1 GDIQ01131261 JAL20465.1 GBHO01034006 GBHO01017924 JAG09598.1 JAG25680.1 GDIP01041878 JAM61837.1 GGFJ01000137 MBW49278.1 GDIP01172109 JAJ51293.1 GDIQ01238178 JAK13547.1 CH954179 KQS62385.1 GDIQ01054388 JAN40349.1 GDIQ01242728 JAK08997.1 GDIQ01062442 JAN32295.1 EDV55669.1 KQS62386.1 KQS62387.1 GGMR01002411 MBY15030.1 GDIP01096372 JAM07343.1 GDIP01097624 JAM06091.1 GDIP01230563 JAI92838.1 GDIQ01035642 JAN59095.1 GDIP01053087 JAM50628.1 GDIQ01116265 JAL35461.1 GDIQ01088271 JAN06466.1 GDIP01165860 JAJ57542.1 GDIQ01027900 JAN66837.1 GDIP01164863 JAJ58539.1 GDIP01166361 JAJ57041.1 GDIP01147169 JAJ76233.1 GDIQ01031996 JAN62741.1 GDIP01208907 JAJ14495.1 GDIP01185562 JAJ37840.1 GDIQ01130058 JAL21668.1 GDIP01015011 JAM88704.1 GGFK01000632 MBW33953.1 GDIP01161264 JAJ62138.1 GDIP01051933 JAM51782.1 AE013599 AAM68489.1 AAM68490.2 GDIP01062225 JAM41490.1 GDIP01178166 JAJ45236.1 GDIP01185563 JAJ37839.1 GDIP01053088 JAM50627.1 AY069804 AAL39949.1 GDIP01064785 JAM38930.1 GDIP01197012 JAJ26390.1 GDIP01146066 JAJ77336.1 GDIP01113573 JAL90141.1 AFH08134.1 AF032870 AAC38820.1 GDIP01088275 JAM15440.1
GEDC01026077 JAS11221.1 GEDC01025301 JAS11997.1 GEDC01030180 JAS07118.1 GEBQ01013488 JAT26489.1 GEBQ01021782 JAT18195.1 GEBQ01022246 JAT17731.1 GEBQ01001563 JAT38414.1 GECU01012161 JAS95545.1 GFTR01008761 JAW07665.1 GECU01034344 JAS73362.1 NNAY01001923 OXU22552.1 KQ414591 KOC70282.1 KQ797310 OAD46959.1 KQ434879 KZC09940.1 DS235123 EEB12220.1 GBYB01014566 JAG84333.1 GBYB01014570 JAG84337.1 UFQS01000449 UFQT01000449 SSX04115.1 SSX24480.1 SSX04113.1 SSX24478.1 KQ435794 KOX73980.1 GFXV01000080 MBW11885.1 LRGB01002190 KZS08498.1 GDHC01012173 JAQ06456.1 GGFM01000908 MBW21659.1 ADMH02000286 ETN67114.1 GGFL01006566 MBW70744.1 GDHC01011085 JAQ07544.1 GGFK01002174 MBW35495.1 GGFJ01000710 MBW49851.1 GBHO01017925 JAG25679.1 GBHO01022022 GBHO01017923 JAG21582.1 JAG25681.1 GGFK01001808 MBW35129.1 GDIQ01131260 JAL20466.1 GDIQ01065438 JAN29299.1 GDIQ01062441 JAN32296.1 GBHO01034007 JAG09597.1 GGFL01001957 MBW66135.1 GDIQ01131261 JAL20465.1 GBHO01034006 GBHO01017924 JAG09598.1 JAG25680.1 GDIP01041878 JAM61837.1 GGFJ01000137 MBW49278.1 GDIP01172109 JAJ51293.1 GDIQ01238178 JAK13547.1 CH954179 KQS62385.1 GDIQ01054388 JAN40349.1 GDIQ01242728 JAK08997.1 GDIQ01062442 JAN32295.1 EDV55669.1 KQS62386.1 KQS62387.1 GGMR01002411 MBY15030.1 GDIP01096372 JAM07343.1 GDIP01097624 JAM06091.1 GDIP01230563 JAI92838.1 GDIQ01035642 JAN59095.1 GDIP01053087 JAM50628.1 GDIQ01116265 JAL35461.1 GDIQ01088271 JAN06466.1 GDIP01165860 JAJ57542.1 GDIQ01027900 JAN66837.1 GDIP01164863 JAJ58539.1 GDIP01166361 JAJ57041.1 GDIP01147169 JAJ76233.1 GDIQ01031996 JAN62741.1 GDIP01208907 JAJ14495.1 GDIP01185562 JAJ37840.1 GDIQ01130058 JAL21668.1 GDIP01015011 JAM88704.1 GGFK01000632 MBW33953.1 GDIP01161264 JAJ62138.1 GDIP01051933 JAM51782.1 AE013599 AAM68489.1 AAM68490.2 GDIP01062225 JAM41490.1 GDIP01178166 JAJ45236.1 GDIP01185563 JAJ37839.1 GDIP01053088 JAM50627.1 AY069804 AAL39949.1 GDIP01064785 JAM38930.1 GDIP01197012 JAJ26390.1 GDIP01146066 JAJ77336.1 GDIP01113573 JAL90141.1 AFH08134.1 AF032870 AAC38820.1 GDIP01088275 JAM15440.1
Proteomes
Pfam
Interpro
IPR002219
PE/DAG-bd
+ More
IPR015212 RGS-like_dom
IPR041489 PDZ_6
IPR036305 RGS_sf
IPR036034 PDZ_sf
IPR001478 PDZ
IPR020454 DAG/PE-bd
IPR016137 RGS
IPR000219 DH-domain
IPR001849 PH_domain
IPR035899 DBL_dom_sf
IPR041020 PH_16
IPR011993 PH-like_dom_sf
IPR011677 DUF1619
IPR010796 B9_dom
IPR005822 Ribosomal_L13
IPR036899 Ribosomal_L13_sf
IPR015212 RGS-like_dom
IPR041489 PDZ_6
IPR036305 RGS_sf
IPR036034 PDZ_sf
IPR001478 PDZ
IPR020454 DAG/PE-bd
IPR016137 RGS
IPR000219 DH-domain
IPR001849 PH_domain
IPR035899 DBL_dom_sf
IPR041020 PH_16
IPR011993 PH-like_dom_sf
IPR011677 DUF1619
IPR010796 B9_dom
IPR005822 Ribosomal_L13
IPR036899 Ribosomal_L13_sf
Gene 3D
ProteinModelPortal
H9JF58
A0A1B6JC30
A0A1B6DCQ4
A0A1B6CCT4
A0A1B6CEY4
A0A1B6C0U4
+ More
A0A1B6LSD5 A0A1B6L3G5 A0A1B6L264 A0A1B6MR53 A0A1B6J8S6 A0A224X591 A0A1B6HFE4 A0A232EW19 K7JHC5 A0A0L7RH29 A0A310SED8 A0A154PFD2 E0VFR4 A0A0C9S1B8 A0A0C9RYX7 A0A336KLF3 A0A336MEI9 A0A0M8ZYZ7 A0A2H8TCQ7 A0A164RBB5 A0A1W4UZR7 A0A1W4VDC9 A0A146LHP2 A0A2M3YZF4 A0A1W4VC14 W5JW08 A0A2M4CZJ0 A0A146LHC1 A0A2M4A418 A0A2M4BA38 A0A0A9Y0I0 A0A0A9Y211 A0A2M4A2Y9 A0A0P5PQ09 A0A0P6FDE1 A0A0P6EL40 A0A0A9WME6 A0A2M4CLI9 A0A0P5PG33 A0A0A9Y833 A0A182FVX8 A0A0P5ZLI1 A0A2M4B8X3 A0A0P5CMD4 A0A0P5IH19 A0A0Q5VM08 A0A0N8E7G0 A0A0P5GD22 A0A0P6EHW8 B3NP30 A0A2S2NCX6 A0A0P5VRN3 A0A0P5VNJ4 A0A0P4Y9Z7 A0A0P6H2V5 A0A0N8DAE5 A0A0P5QUN2 A0A0P6CRF7 A0A0P5CTM7 A0A0N8EGX1 A0A0P5CW19 A0A0N8AAJ7 A0A0N8AHE6 A0A0N8EFF9 A0A0P4ZT61 A0A0P5BF15 A0A0P5PDU4 A0A0P6C0W5 A0A2M3ZZH5 A0A0P5D8V5 A0A0P5YXE7 A1ZAN6 A0A0P5YAW3 A0A0P5BYH5 A0A0P5BF40 A0A0P5YV04 Q8T9E3 A0A0P5Y2W1 A0A0P5ALS5 A0A0P5E7C4 A0A0P5UPY8 A0A0B4K7T7 O44381 A0A0P5WDP8
A0A1B6LSD5 A0A1B6L3G5 A0A1B6L264 A0A1B6MR53 A0A1B6J8S6 A0A224X591 A0A1B6HFE4 A0A232EW19 K7JHC5 A0A0L7RH29 A0A310SED8 A0A154PFD2 E0VFR4 A0A0C9S1B8 A0A0C9RYX7 A0A336KLF3 A0A336MEI9 A0A0M8ZYZ7 A0A2H8TCQ7 A0A164RBB5 A0A1W4UZR7 A0A1W4VDC9 A0A146LHP2 A0A2M3YZF4 A0A1W4VC14 W5JW08 A0A2M4CZJ0 A0A146LHC1 A0A2M4A418 A0A2M4BA38 A0A0A9Y0I0 A0A0A9Y211 A0A2M4A2Y9 A0A0P5PQ09 A0A0P6FDE1 A0A0P6EL40 A0A0A9WME6 A0A2M4CLI9 A0A0P5PG33 A0A0A9Y833 A0A182FVX8 A0A0P5ZLI1 A0A2M4B8X3 A0A0P5CMD4 A0A0P5IH19 A0A0Q5VM08 A0A0N8E7G0 A0A0P5GD22 A0A0P6EHW8 B3NP30 A0A2S2NCX6 A0A0P5VRN3 A0A0P5VNJ4 A0A0P4Y9Z7 A0A0P6H2V5 A0A0N8DAE5 A0A0P5QUN2 A0A0P6CRF7 A0A0P5CTM7 A0A0N8EGX1 A0A0P5CW19 A0A0N8AAJ7 A0A0N8AHE6 A0A0N8EFF9 A0A0P4ZT61 A0A0P5BF15 A0A0P5PDU4 A0A0P6C0W5 A0A2M3ZZH5 A0A0P5D8V5 A0A0P5YXE7 A1ZAN6 A0A0P5YAW3 A0A0P5BYH5 A0A0P5BF40 A0A0P5YV04 Q8T9E3 A0A0P5Y2W1 A0A0P5ALS5 A0A0P5E7C4 A0A0P5UPY8 A0A0B4K7T7 O44381 A0A0P5WDP8
PDB
3CX8
E-value=1.00616e-06,
Score=126
Ontologies
GO
GO:0005737
GO:0046872
GO:0035556
GO:0005089
GO:0035023
GO:0005840
GO:0003735
GO:0006412
GO:0008360
GO:0007349
GO:0030589
GO:0007374
GO:0035277
GO:0110072
GO:0009611
GO:0016331
GO:0005826
GO:0031532
GO:0070252
GO:0007370
GO:0007377
GO:0050770
GO:0007277
GO:0043519
GO:0030478
GO:0045177
GO:0007375
GO:0045178
GO:0007015
GO:0016476
GO:0035025
GO:0016328
GO:0045179
GO:0090254
GO:0016324
GO:0031461
GO:0016791
GO:0005488
GO:0005507
GO:0004713
GO:0016491
GO:0015930
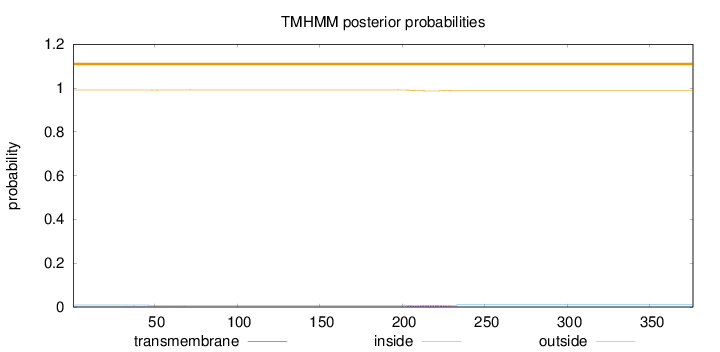
Topology
Length:
376
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1857
Exp number, first 60 AAs:
0.01628
Total prob of N-in:
0.00915
outside
1 - 376
Population Genetic Test Statistics
Pi
320.640828
Theta
197.064404
Tajima's D
2.902016
CLR
0.206112
CSRT
0.974851257437128
Interpretation
Uncertain
