Gene
KWMTBOMO14265
Pre Gene Modal
BGIBMGA008160
Annotation
Tetraspanin_[Operophtera_brumata]
Full name
Tetraspanin
Location in the cell
PlasmaMembrane Reliability : 4.557
Sequence
CDS
ATGACTTCGTTGTGTCTCACGGTCTTCCTGTGCGCATTCAATGTCATACAAGTGCTGTCCGCGTCAGTCCTCGGTGGTGTAGCGATATGGTCCTTCATTGAGGTCGTGAGCCTGACCTCCCTGCGCAACCCCATCCATTACCAGCTGAAGCAGACGTTGAACTGGCCCGAGGTGCTCCCATGGGTCTTTCTCATGGCGGCCATTTTGACCATACTGACCGACATATGTGGCTTGTGCGGTTCGAAGAGGAAGAGTCGACCTCTGATTTTGCTGTACGTGGTACTTCAAATGATGTCCATATTGTTGCTGGTTGGTGCTCCTCTCGTCGCCCTTACCCTCGCAGAGGGACCTAACACCGCAGAATTCTTCAAAAGCACAATGTGGGACGTCTTCTACACTAGTATAGACAGCCAAGATGGCGCGTACAATTCCTTCGAAGAGCGGTTGCGATGCTGCGGCGTCAGTAGTCCCAGAGACTATGTGCCAAAAAGGAACAACTTCCCTGCATCTTGCTGCTATTCGGGTAAGCCCTGTGATTTCTCAGATCATTTCGCGAACAAATTGAGCGGATGCAACGGCGTTGTGGACAAATACACCGAATATTTCATCAAAGGAATGTCTGTGGGATCGGTCGTGGTGGCGATCGTTGGCGTAATTGGGTTTTTGATTGCCGTGGTCTTGTCCATGAAGATGAAGACCAAGAAATTGGAACCTGCCGTCGAAACCGAAGCTGATTGTCAGAAAGTGTTGTTGTAA
Protein
MTSLCLTVFLCAFNVIQVLSASVLGGVAIWSFIEVVSLTSLRNPIHYQLKQTLNWPEVLPWVFLMAAILTILTDICGLCGSKRKSRPLILLYVVLQMMSILLLVGAPLVALTLAEGPNTAEFFKSTMWDVFYTSIDSQDGAYNSFEERLRCCGVSSPRDYVPKRNNFPASCCYSGKPCDFSDHFANKLSGCNGVVDKYTEYFIKGMSVGSVVVAIVGVIGFLIAVVLSMKMKTKKLEPAVETEADCQKVLL
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Feature
chain Tetraspanin
Uniprot
H9JF63
H9JF64
A0A2H1WQL1
A0A0L7LID5
A0A194Q8C3
A0A0N0PCZ3
+ More
A0A2A4JVV7 A0A2W1BL09 A0A212FJD9 H9JF65 B2DBM8 A0A0N0PD86 A0A2W1BKX8 A0A2A4JV62 S4PCJ6 A0A2H1WQP0 A0A212FJB6 A0A0L7LIZ1 A0A1E1W0A5 A0A1Y1L5M1 A0A315UWP8 A0A151I4G3 H3CW26 A0A3B5KLI5 A0A1L8DK44 A0A147A3D7 A0A1L8DK45 Q4SHG2 A0A3S2N5Y2 Q1LWF8 H2UYH5 Q6P031 A0A146WIY6 A0A3B4BBC3 A0A3B3ULM4 A0A3P9PCA8 W8ATQ8 A0A3B3BHQ0 A0A3B3ULN6 M3ZCS7 A0A3P9N9M2 A0A3P9N9K2 A0A151JNQ2 A0A315WBC6 A0A3B5MBA1 A0A3B5RBC2 A0A3B3YJ36 A0A087XCU0 A0A147A2Y1 A0A3B4ZYP6 A0A0S7HNP1 A0A0C9R0W0 A0A3Q1JAK1 A0A3B3S7P0 A0A146PKL7 A0A0S7LJQ2 A0A131ZWC3 A0A0C9R490 A0A0P5U5V8 A0A139WAJ4 A0A0C9QLV9 A0A0C9Q5P6 A0A0P6CK49 A0A3Q0STD0 A0A1W5B0K8 A0A0T6B8H7 D3PG37 A0A3Q2GJ42 A0A0K2TX70 A0A0L0C071 A0A087XCV8 A0A3Q1GMK1 Q16HS3 A0A0P4WC72 A0A0P6IJZ2 A0A3B5MHM8 A0A3Q4AFS0 A0A3B4X0R8 A0A3Q1AGY3 A0A3P8RJ81 M4A2U9 A0A3Q1BXJ2 A0A3B4UPQ0 E0VNB3 A0A224XM16 U6J0I8 J3S154 A0A3B4UNV9 A0A068Y7H8 I3JEP3
A0A2A4JVV7 A0A2W1BL09 A0A212FJD9 H9JF65 B2DBM8 A0A0N0PD86 A0A2W1BKX8 A0A2A4JV62 S4PCJ6 A0A2H1WQP0 A0A212FJB6 A0A0L7LIZ1 A0A1E1W0A5 A0A1Y1L5M1 A0A315UWP8 A0A151I4G3 H3CW26 A0A3B5KLI5 A0A1L8DK44 A0A147A3D7 A0A1L8DK45 Q4SHG2 A0A3S2N5Y2 Q1LWF8 H2UYH5 Q6P031 A0A146WIY6 A0A3B4BBC3 A0A3B3ULM4 A0A3P9PCA8 W8ATQ8 A0A3B3BHQ0 A0A3B3ULN6 M3ZCS7 A0A3P9N9M2 A0A3P9N9K2 A0A151JNQ2 A0A315WBC6 A0A3B5MBA1 A0A3B5RBC2 A0A3B3YJ36 A0A087XCU0 A0A147A2Y1 A0A3B4ZYP6 A0A0S7HNP1 A0A0C9R0W0 A0A3Q1JAK1 A0A3B3S7P0 A0A146PKL7 A0A0S7LJQ2 A0A131ZWC3 A0A0C9R490 A0A0P5U5V8 A0A139WAJ4 A0A0C9QLV9 A0A0C9Q5P6 A0A0P6CK49 A0A3Q0STD0 A0A1W5B0K8 A0A0T6B8H7 D3PG37 A0A3Q2GJ42 A0A0K2TX70 A0A0L0C071 A0A087XCV8 A0A3Q1GMK1 Q16HS3 A0A0P4WC72 A0A0P6IJZ2 A0A3B5MHM8 A0A3Q4AFS0 A0A3B4X0R8 A0A3Q1AGY3 A0A3P8RJ81 M4A2U9 A0A3Q1BXJ2 A0A3B4UPQ0 E0VNB3 A0A224XM16 U6J0I8 J3S154 A0A3B4UNV9 A0A068Y7H8 I3JEP3
Pubmed
EMBL
BABH01026802
ODYU01010303
SOQ55318.1
JTDY01001003
KOB75145.1
KQ459386
+ More
KPJ01240.1 KQ460423 KPJ14817.1 NWSH01000516 PCG75916.1 KZ150056 PZC74324.1 AGBW02008277 OWR53846.1 AB264712 BAG30787.1 KPJ01239.1 KPJ14818.1 PZC74325.1 PCG75915.1 GAIX01005382 JAA87178.1 SOQ55317.1 OWR53847.1 KOB75146.1 GDQN01010637 GDQN01002817 JAT80417.1 JAT88237.1 GEZM01064003 JAV68874.1 NHOQ01002686 PWA15541.1 KQ976462 KYM84644.1 GFDF01007252 JAV06832.1 GCES01013252 JAR73071.1 GFDF01007251 JAV06833.1 CAAE01014581 CAF99920.1 CM012439 RVE74216.1 CU656039 FO834814 BC065859 AAH65859.1 GCES01057346 JAR28977.1 GAMC01018522 JAB88033.1 KQ978844 KYN27991.1 NHOQ01000682 PWA29156.1 AYCK01012806 GCES01013390 JAR72933.1 GBYX01438561 JAO42792.1 GBYB01001630 JAG71397.1 GCES01142047 JAQ44275.1 GBYX01165500 GBYX01165498 JAO89687.1 JXLN01004310 KPM03168.1 GBYB01001631 JAG71398.1 GDIP01134610 LRGB01002559 JAL69104.1 KZS07150.1 KQ971381 KYB24952.1 GBYB01001632 JAG71399.1 GBYB01009418 JAG79185.1 GDIP01007029 JAM96686.1 LJIG01009211 KRT83534.1 BT120593 ADD24233.1 HACA01012936 CDW30297.1 JRES01001077 KNC25695.1 AYCK01017622 CH478145 EAT33803.1 GDRN01082066 JAI61911.1 GDIQ01020589 JAN74148.1 DS235335 EEB14869.1 GFTR01004292 JAW12134.1 LK028576 APAU02000005 CDS15929.1 EUB63766.1 JU176095 AFJ51619.1 LN902841 CDS40740.1 AERX01024909
KPJ01240.1 KQ460423 KPJ14817.1 NWSH01000516 PCG75916.1 KZ150056 PZC74324.1 AGBW02008277 OWR53846.1 AB264712 BAG30787.1 KPJ01239.1 KPJ14818.1 PZC74325.1 PCG75915.1 GAIX01005382 JAA87178.1 SOQ55317.1 OWR53847.1 KOB75146.1 GDQN01010637 GDQN01002817 JAT80417.1 JAT88237.1 GEZM01064003 JAV68874.1 NHOQ01002686 PWA15541.1 KQ976462 KYM84644.1 GFDF01007252 JAV06832.1 GCES01013252 JAR73071.1 GFDF01007251 JAV06833.1 CAAE01014581 CAF99920.1 CM012439 RVE74216.1 CU656039 FO834814 BC065859 AAH65859.1 GCES01057346 JAR28977.1 GAMC01018522 JAB88033.1 KQ978844 KYN27991.1 NHOQ01000682 PWA29156.1 AYCK01012806 GCES01013390 JAR72933.1 GBYX01438561 JAO42792.1 GBYB01001630 JAG71397.1 GCES01142047 JAQ44275.1 GBYX01165500 GBYX01165498 JAO89687.1 JXLN01004310 KPM03168.1 GBYB01001631 JAG71398.1 GDIP01134610 LRGB01002559 JAL69104.1 KZS07150.1 KQ971381 KYB24952.1 GBYB01001632 JAG71399.1 GBYB01009418 JAG79185.1 GDIP01007029 JAM96686.1 LJIG01009211 KRT83534.1 BT120593 ADD24233.1 HACA01012936 CDW30297.1 JRES01001077 KNC25695.1 AYCK01017622 CH478145 EAT33803.1 GDRN01082066 JAI61911.1 GDIQ01020589 JAN74148.1 DS235335 EEB14869.1 GFTR01004292 JAW12134.1 LK028576 APAU02000005 CDS15929.1 EUB63766.1 JU176095 AFJ51619.1 LN902841 CDS40740.1 AERX01024909
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000218220
UP000007151
+ More
UP000078540 UP000007303 UP000005226 UP000000437 UP000261520 UP000261500 UP000242638 UP000261560 UP000002852 UP000078492 UP000261380 UP000261480 UP000028760 UP000261400 UP000265040 UP000261540 UP000076858 UP000007266 UP000261340 UP000192224 UP000265020 UP000037069 UP000257200 UP000008820 UP000261620 UP000261360 UP000257160 UP000265080 UP000261420 UP000009046 UP000019149 UP000017246 UP000005207
UP000078540 UP000007303 UP000005226 UP000000437 UP000261520 UP000261500 UP000242638 UP000261560 UP000002852 UP000078492 UP000261380 UP000261480 UP000028760 UP000261400 UP000265040 UP000261540 UP000076858 UP000007266 UP000261340 UP000192224 UP000265020 UP000037069 UP000257200 UP000008820 UP000261620 UP000261360 UP000257160 UP000265080 UP000261420 UP000009046 UP000019149 UP000017246 UP000005207
Pfam
PF00335 Tetraspanin
Interpro
SUPFAM
SSF48652
SSF48652
Gene 3D
ProteinModelPortal
H9JF63
H9JF64
A0A2H1WQL1
A0A0L7LID5
A0A194Q8C3
A0A0N0PCZ3
+ More
A0A2A4JVV7 A0A2W1BL09 A0A212FJD9 H9JF65 B2DBM8 A0A0N0PD86 A0A2W1BKX8 A0A2A4JV62 S4PCJ6 A0A2H1WQP0 A0A212FJB6 A0A0L7LIZ1 A0A1E1W0A5 A0A1Y1L5M1 A0A315UWP8 A0A151I4G3 H3CW26 A0A3B5KLI5 A0A1L8DK44 A0A147A3D7 A0A1L8DK45 Q4SHG2 A0A3S2N5Y2 Q1LWF8 H2UYH5 Q6P031 A0A146WIY6 A0A3B4BBC3 A0A3B3ULM4 A0A3P9PCA8 W8ATQ8 A0A3B3BHQ0 A0A3B3ULN6 M3ZCS7 A0A3P9N9M2 A0A3P9N9K2 A0A151JNQ2 A0A315WBC6 A0A3B5MBA1 A0A3B5RBC2 A0A3B3YJ36 A0A087XCU0 A0A147A2Y1 A0A3B4ZYP6 A0A0S7HNP1 A0A0C9R0W0 A0A3Q1JAK1 A0A3B3S7P0 A0A146PKL7 A0A0S7LJQ2 A0A131ZWC3 A0A0C9R490 A0A0P5U5V8 A0A139WAJ4 A0A0C9QLV9 A0A0C9Q5P6 A0A0P6CK49 A0A3Q0STD0 A0A1W5B0K8 A0A0T6B8H7 D3PG37 A0A3Q2GJ42 A0A0K2TX70 A0A0L0C071 A0A087XCV8 A0A3Q1GMK1 Q16HS3 A0A0P4WC72 A0A0P6IJZ2 A0A3B5MHM8 A0A3Q4AFS0 A0A3B4X0R8 A0A3Q1AGY3 A0A3P8RJ81 M4A2U9 A0A3Q1BXJ2 A0A3B4UPQ0 E0VNB3 A0A224XM16 U6J0I8 J3S154 A0A3B4UNV9 A0A068Y7H8 I3JEP3
A0A2A4JVV7 A0A2W1BL09 A0A212FJD9 H9JF65 B2DBM8 A0A0N0PD86 A0A2W1BKX8 A0A2A4JV62 S4PCJ6 A0A2H1WQP0 A0A212FJB6 A0A0L7LIZ1 A0A1E1W0A5 A0A1Y1L5M1 A0A315UWP8 A0A151I4G3 H3CW26 A0A3B5KLI5 A0A1L8DK44 A0A147A3D7 A0A1L8DK45 Q4SHG2 A0A3S2N5Y2 Q1LWF8 H2UYH5 Q6P031 A0A146WIY6 A0A3B4BBC3 A0A3B3ULM4 A0A3P9PCA8 W8ATQ8 A0A3B3BHQ0 A0A3B3ULN6 M3ZCS7 A0A3P9N9M2 A0A3P9N9K2 A0A151JNQ2 A0A315WBC6 A0A3B5MBA1 A0A3B5RBC2 A0A3B3YJ36 A0A087XCU0 A0A147A2Y1 A0A3B4ZYP6 A0A0S7HNP1 A0A0C9R0W0 A0A3Q1JAK1 A0A3B3S7P0 A0A146PKL7 A0A0S7LJQ2 A0A131ZWC3 A0A0C9R490 A0A0P5U5V8 A0A139WAJ4 A0A0C9QLV9 A0A0C9Q5P6 A0A0P6CK49 A0A3Q0STD0 A0A1W5B0K8 A0A0T6B8H7 D3PG37 A0A3Q2GJ42 A0A0K2TX70 A0A0L0C071 A0A087XCV8 A0A3Q1GMK1 Q16HS3 A0A0P4WC72 A0A0P6IJZ2 A0A3B5MHM8 A0A3Q4AFS0 A0A3B4X0R8 A0A3Q1AGY3 A0A3P8RJ81 M4A2U9 A0A3Q1BXJ2 A0A3B4UPQ0 E0VNB3 A0A224XM16 U6J0I8 J3S154 A0A3B4UNV9 A0A068Y7H8 I3JEP3
Ontologies
GO
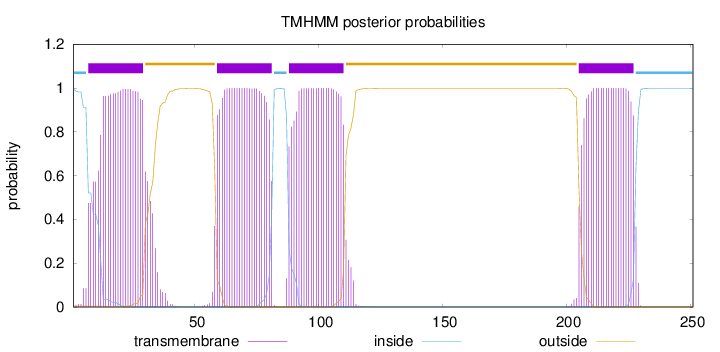
Topology
Subcellular location
Membrane
Length:
251
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
90.97235
Exp number, first 60 AAs:
25.39574
Total prob of N-in:
0.99543
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 58
TMhelix
59 - 81
inside
82 - 87
TMhelix
88 - 110
outside
111 - 204
TMhelix
205 - 227
inside
228 - 251
Population Genetic Test Statistics
Pi
389.498818
Theta
234.76371
Tajima's D
2.07983
CLR
0
CSRT
0.892405379731013
Interpretation
Uncertain
