Gene
KWMTBOMO14263 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008162
Annotation
PREDICTED:_CD151_antigen-like_[Papilio_polytes]
Full name
Tetraspanin
Location in the cell
PlasmaMembrane Reliability : 4.832
Sequence
CDS
ATGTCCAAGGAGGAACTTATCAAAGGAGTGTTGTACGTTGTCAATGTGATCTACGCTGTTTTCGGTCTAGTCACCGCCGCGACCGGGATATGGTTCTTTGTGCAATTAGCCGAGTTCGTCAGTCTCAGAAACAGCAACCATTACTTGCTGGACTACCGCGTCTACTGGCCACAGGTCGCGCCATGGCTCTTCATCCTGCTGGGTCTGTTTGTTATGATGGTGGCGTTTTGCGGATGGTGCGGCGCTAACAAAGAGAGCCGAGGACTTCTGGGCATCTACGGGATCTTCCTGATCCTCATCATCGTTGGACAAGCGATCGCAGCCACGTTAATCTTCGTGTTTGTGGACGGAGAAGACACTGACCGCTTCATCAAGGACACTGTCTACGACGGCTACTACAACTCCCAGTCGAATCCTGACGTGTTCAAAGCGTTCGGGAGGATTGAAAGGAAACTGAGGTGTTGCGGTGCGAACGACGCCAGGGACTATCGGAGCTGGAGAAACGATCTGCCCCTGACGTGCTGCCTGGACAGCTACTACCGGGCCTCCTGCGACTTCACCGACAAGGAAGCCAACGAGCGTCTCGGCTGTGCCAAGGTCGCATCCGTGTACACCAAAATCATCAGCTCTTCGGTCGCCGGAGCGTCGCTTCTGATATCGCTAGTGGAAATAGCAGGCGTCGTGCTGGCCTGGAGACTGTTTCACTCGCTGCGAGAGGTGGAACACTACATCCATGAGCGGAAGGAAGGTGAAACCGAATGCTAG
Protein
MSKEELIKGVLYVVNVIYAVFGLVTAATGIWFFVQLAEFVSLRNSNHYLLDYRVYWPQVAPWLFILLGLFVMMVAFCGWCGANKESRGLLGIYGIFLILIIVGQAIAATLIFVFVDGEDTDRFIKDTVYDGYYNSQSNPDVFKAFGRIERKLRCCGANDARDYRSWRNDLPLTCCLDSYYRASCDFTDKEANERLGCAKVASVYTKIISSSVAGASLLISLVEIAGVVLAWRLFHSLREVEHYIHERKEGETEC
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Feature
chain Tetraspanin
Uniprot
H9JF65
A0A212FJD9
S4PCJ6
A0A2W1BKX8
A0A2A4JV62
A0A0L7LIZ1
+ More
A0A2H1WQP0 B2DBM8 A0A0N0PD86 A0A212FJB6 A0A0L7LID5 A0A2A4JVV7 A0A2W1BL09 A0A0N0PCZ3 A0A194Q8C3 H9JF64 A0A2H1WQL1 H9JF63 A0A1E1W0A5 A0A1E1WV10 A0A1E1W4P8 S4PWL6 I4DMY0 A0A087TFG6 A0A212EWY8 R4UW62 A0A212F0C8 A0A087ZPN3 I4DJ82 A0A232FLJ2 K7IST2 Q9NB07 A0A139WAJ4 A0A2A3EHK8 V9IIY1 A0A3B3BHQ0 Q9NB08 T1IXU1 A0A158NAZ0 S4PYD2 A0A0M5J749 D3PG37 A0A2T7PKV1 A0A158NJM3 A0A1Z5L5C1 A0A194Q3K1 A0A0K2TX70 H9H6R1 A0A3M6V1N1 A0A1I8QDE5 A0A2A4IYD2 A0A1U7Q6J3 A0A3Q1JUX4 A0A2R5L585 A0A3L8DMA2 E2A971 B4MQJ7 A0A067QFM7 A0A061IBK7 A0A3B3YJ36 A0A087XCU0 A0A3B4T8X4 B1WAT5 A0A0S7LJQ2 A0A3B4XQ94 A0A0J7L311 U5EVW1 A0A3B4X0R8 E0VTU3 C3Y656 A0A067QYE5 K7J9H7 D2A3F2 A0A383Z5F2 A0A0A0V5H3 V5I9N6 A0A3B3Q3F5 Q6DDI3 A0A1L8GCX7 A0A383Z5B6 A0A2B4SAC1 A0A2A4K689 K1R7F9 A0A023EK84 W8ATQ8 A0A293MV59 A0A1Y1L5M1 M3ZCS7 T1E2B6 E2BYU7 A0A1W5B0K8 A0A3S2N5Y2 A0A1I8PKV3 A0A3Q1BMI7 A0A0L0CN39 A0A3P9N9K2 A0A3S1CB08 A0A076KUR5 A0A023EK90
A0A2H1WQP0 B2DBM8 A0A0N0PD86 A0A212FJB6 A0A0L7LID5 A0A2A4JVV7 A0A2W1BL09 A0A0N0PCZ3 A0A194Q8C3 H9JF64 A0A2H1WQL1 H9JF63 A0A1E1W0A5 A0A1E1WV10 A0A1E1W4P8 S4PWL6 I4DMY0 A0A087TFG6 A0A212EWY8 R4UW62 A0A212F0C8 A0A087ZPN3 I4DJ82 A0A232FLJ2 K7IST2 Q9NB07 A0A139WAJ4 A0A2A3EHK8 V9IIY1 A0A3B3BHQ0 Q9NB08 T1IXU1 A0A158NAZ0 S4PYD2 A0A0M5J749 D3PG37 A0A2T7PKV1 A0A158NJM3 A0A1Z5L5C1 A0A194Q3K1 A0A0K2TX70 H9H6R1 A0A3M6V1N1 A0A1I8QDE5 A0A2A4IYD2 A0A1U7Q6J3 A0A3Q1JUX4 A0A2R5L585 A0A3L8DMA2 E2A971 B4MQJ7 A0A067QFM7 A0A061IBK7 A0A3B3YJ36 A0A087XCU0 A0A3B4T8X4 B1WAT5 A0A0S7LJQ2 A0A3B4XQ94 A0A0J7L311 U5EVW1 A0A3B4X0R8 E0VTU3 C3Y656 A0A067QYE5 K7J9H7 D2A3F2 A0A383Z5F2 A0A0A0V5H3 V5I9N6 A0A3B3Q3F5 Q6DDI3 A0A1L8GCX7 A0A383Z5B6 A0A2B4SAC1 A0A2A4K689 K1R7F9 A0A023EK84 W8ATQ8 A0A293MV59 A0A1Y1L5M1 M3ZCS7 T1E2B6 E2BYU7 A0A1W5B0K8 A0A3S2N5Y2 A0A1I8PKV3 A0A3Q1BMI7 A0A0L0CN39 A0A3P9N9K2 A0A3S1CB08 A0A076KUR5 A0A023EK90
Pubmed
19121390
22118469
23622113
28756777
26227816
18712529
+ More
26354079 22651552 28648823 20075255 11122467 18362917 19820115 29451363 21347285 28528879 17495919 30382153 30249741 20798317 17994087 24845553 23929341 29704459 20431018 20566863 18563158 24303891 29240929 27762356 22992520 24945155 24495485 28004739 23542700 24330624 26108605
26354079 22651552 28648823 20075255 11122467 18362917 19820115 29451363 21347285 28528879 17495919 30382153 30249741 20798317 17994087 24845553 23929341 29704459 20431018 20566863 18563158 24303891 29240929 27762356 22992520 24945155 24495485 28004739 23542700 24330624 26108605
EMBL
BABH01026802
AGBW02008277
OWR53846.1
GAIX01005382
JAA87178.1
KZ150056
+ More
PZC74325.1 NWSH01000516 PCG75915.1 JTDY01001003 KOB75146.1 ODYU01010303 SOQ55317.1 AB264712 KQ459386 BAG30787.1 KPJ01239.1 KQ460423 KPJ14818.1 OWR53847.1 KOB75145.1 PCG75916.1 PZC74324.1 KPJ14817.1 KPJ01240.1 SOQ55318.1 GDQN01010637 GDQN01002817 JAT80417.1 JAT88237.1 GDQN01000423 JAT90631.1 GDQN01010381 GDQN01009120 JAT80673.1 JAT81934.1 GAIX01006573 JAA85987.1 AK402648 BAM19270.1 KK114976 KFM63855.1 AGBW02011867 OWR45999.1 KC571982 AGM32481.1 AGBW02011105 OWR47199.1 AK401350 BAM17972.1 NNAY01000043 OXU31611.1 AAZX01003309 AAZX01016448 AF274024 AAF90150.1 KQ971381 KYB24952.1 KZ288246 PBC31188.1 JR049622 AEY61020.1 AF274023 AAF90149.1 JH431663 ADTU01010579 ADTU01010580 ADTU01010581 GAIX01003493 JAA89067.1 CP012524 ALC41286.1 BT120593 ADD24233.1 PZQS01000003 PVD34030.1 ADTU01001946 ADTU01001947 ADTU01001948 GFJQ02004357 JAW02613.1 KQ459472 KPJ00118.1 HACA01012936 CDW30297.1 RCHS01000292 RMX59684.1 NWSH01005565 PCG64123.1 GGLE01000471 MBY04597.1 QOIP01000006 RLU21537.1 GL437711 EFN70056.1 CH963849 EDW74386.1 KK853603 KDR06366.1 KE672658 RAZU01000139 ERE79075.1 RLQ70269.1 AYCK01012806 AAMC01052796 BC161493 AAI61493.1 GBYX01165500 GBYX01165498 JAO89687.1 LBMM01001031 KMQ97046.1 GANO01001692 JAB58179.1 DS235773 EEB16799.1 GG666487 EEN64453.1 KK853109 KDR11258.1 KQ971338 EFA01921.1 KF860450 AIW62442.1 GALX01002620 JAB65846.1 BC077579 CM004472 AAH77579.1 OCT83961.1 CM004473 OCT81779.1 LSMT01000143 PFX25760.1 NWSH01000112 PCG79438.1 JH816125 EKC37090.1 GAPW01004057 JAC09541.1 GAMC01018522 JAB88033.1 GFWV01019995 MAA44723.1 GEZM01064003 JAV68874.1 GALA01000879 JAA93973.1 GL451531 EFN79119.1 CM012439 RVE74216.1 JRES01000160 KNC33725.1 RQTK01000099 RUS87841.1 KF433416 AII97740.1 GAPW01004058 JAC09540.1
PZC74325.1 NWSH01000516 PCG75915.1 JTDY01001003 KOB75146.1 ODYU01010303 SOQ55317.1 AB264712 KQ459386 BAG30787.1 KPJ01239.1 KQ460423 KPJ14818.1 OWR53847.1 KOB75145.1 PCG75916.1 PZC74324.1 KPJ14817.1 KPJ01240.1 SOQ55318.1 GDQN01010637 GDQN01002817 JAT80417.1 JAT88237.1 GDQN01000423 JAT90631.1 GDQN01010381 GDQN01009120 JAT80673.1 JAT81934.1 GAIX01006573 JAA85987.1 AK402648 BAM19270.1 KK114976 KFM63855.1 AGBW02011867 OWR45999.1 KC571982 AGM32481.1 AGBW02011105 OWR47199.1 AK401350 BAM17972.1 NNAY01000043 OXU31611.1 AAZX01003309 AAZX01016448 AF274024 AAF90150.1 KQ971381 KYB24952.1 KZ288246 PBC31188.1 JR049622 AEY61020.1 AF274023 AAF90149.1 JH431663 ADTU01010579 ADTU01010580 ADTU01010581 GAIX01003493 JAA89067.1 CP012524 ALC41286.1 BT120593 ADD24233.1 PZQS01000003 PVD34030.1 ADTU01001946 ADTU01001947 ADTU01001948 GFJQ02004357 JAW02613.1 KQ459472 KPJ00118.1 HACA01012936 CDW30297.1 RCHS01000292 RMX59684.1 NWSH01005565 PCG64123.1 GGLE01000471 MBY04597.1 QOIP01000006 RLU21537.1 GL437711 EFN70056.1 CH963849 EDW74386.1 KK853603 KDR06366.1 KE672658 RAZU01000139 ERE79075.1 RLQ70269.1 AYCK01012806 AAMC01052796 BC161493 AAI61493.1 GBYX01165500 GBYX01165498 JAO89687.1 LBMM01001031 KMQ97046.1 GANO01001692 JAB58179.1 DS235773 EEB16799.1 GG666487 EEN64453.1 KK853109 KDR11258.1 KQ971338 EFA01921.1 KF860450 AIW62442.1 GALX01002620 JAB65846.1 BC077579 CM004472 AAH77579.1 OCT83961.1 CM004473 OCT81779.1 LSMT01000143 PFX25760.1 NWSH01000112 PCG79438.1 JH816125 EKC37090.1 GAPW01004057 JAC09541.1 GAMC01018522 JAB88033.1 GFWV01019995 MAA44723.1 GEZM01064003 JAV68874.1 GALA01000879 JAA93973.1 GL451531 EFN79119.1 CM012439 RVE74216.1 JRES01000160 KNC33725.1 RQTK01000099 RUS87841.1 KF433416 AII97740.1 GAPW01004058 JAC09540.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000037510
UP000053268
UP000053240
+ More
UP000054359 UP000005203 UP000215335 UP000002358 UP000007266 UP000242457 UP000261560 UP000005205 UP000092553 UP000245119 UP000002280 UP000275408 UP000095300 UP000189706 UP000265040 UP000279307 UP000000311 UP000007798 UP000027135 UP000030759 UP000273346 UP000261480 UP000028760 UP000261420 UP000008143 UP000261360 UP000036403 UP000009046 UP000001554 UP000261681 UP000261540 UP000186698 UP000225706 UP000005408 UP000002852 UP000008237 UP000192224 UP000257160 UP000037069 UP000242638 UP000271974
UP000054359 UP000005203 UP000215335 UP000002358 UP000007266 UP000242457 UP000261560 UP000005205 UP000092553 UP000245119 UP000002280 UP000275408 UP000095300 UP000189706 UP000265040 UP000279307 UP000000311 UP000007798 UP000027135 UP000030759 UP000273346 UP000261480 UP000028760 UP000261420 UP000008143 UP000261360 UP000036403 UP000009046 UP000001554 UP000261681 UP000261540 UP000186698 UP000225706 UP000005408 UP000002852 UP000008237 UP000192224 UP000257160 UP000037069 UP000242638 UP000271974
Pfam
PF00335 Tetraspanin
Interpro
Gene 3D
ProteinModelPortal
H9JF65
A0A212FJD9
S4PCJ6
A0A2W1BKX8
A0A2A4JV62
A0A0L7LIZ1
+ More
A0A2H1WQP0 B2DBM8 A0A0N0PD86 A0A212FJB6 A0A0L7LID5 A0A2A4JVV7 A0A2W1BL09 A0A0N0PCZ3 A0A194Q8C3 H9JF64 A0A2H1WQL1 H9JF63 A0A1E1W0A5 A0A1E1WV10 A0A1E1W4P8 S4PWL6 I4DMY0 A0A087TFG6 A0A212EWY8 R4UW62 A0A212F0C8 A0A087ZPN3 I4DJ82 A0A232FLJ2 K7IST2 Q9NB07 A0A139WAJ4 A0A2A3EHK8 V9IIY1 A0A3B3BHQ0 Q9NB08 T1IXU1 A0A158NAZ0 S4PYD2 A0A0M5J749 D3PG37 A0A2T7PKV1 A0A158NJM3 A0A1Z5L5C1 A0A194Q3K1 A0A0K2TX70 H9H6R1 A0A3M6V1N1 A0A1I8QDE5 A0A2A4IYD2 A0A1U7Q6J3 A0A3Q1JUX4 A0A2R5L585 A0A3L8DMA2 E2A971 B4MQJ7 A0A067QFM7 A0A061IBK7 A0A3B3YJ36 A0A087XCU0 A0A3B4T8X4 B1WAT5 A0A0S7LJQ2 A0A3B4XQ94 A0A0J7L311 U5EVW1 A0A3B4X0R8 E0VTU3 C3Y656 A0A067QYE5 K7J9H7 D2A3F2 A0A383Z5F2 A0A0A0V5H3 V5I9N6 A0A3B3Q3F5 Q6DDI3 A0A1L8GCX7 A0A383Z5B6 A0A2B4SAC1 A0A2A4K689 K1R7F9 A0A023EK84 W8ATQ8 A0A293MV59 A0A1Y1L5M1 M3ZCS7 T1E2B6 E2BYU7 A0A1W5B0K8 A0A3S2N5Y2 A0A1I8PKV3 A0A3Q1BMI7 A0A0L0CN39 A0A3P9N9K2 A0A3S1CB08 A0A076KUR5 A0A023EK90
A0A2H1WQP0 B2DBM8 A0A0N0PD86 A0A212FJB6 A0A0L7LID5 A0A2A4JVV7 A0A2W1BL09 A0A0N0PCZ3 A0A194Q8C3 H9JF64 A0A2H1WQL1 H9JF63 A0A1E1W0A5 A0A1E1WV10 A0A1E1W4P8 S4PWL6 I4DMY0 A0A087TFG6 A0A212EWY8 R4UW62 A0A212F0C8 A0A087ZPN3 I4DJ82 A0A232FLJ2 K7IST2 Q9NB07 A0A139WAJ4 A0A2A3EHK8 V9IIY1 A0A3B3BHQ0 Q9NB08 T1IXU1 A0A158NAZ0 S4PYD2 A0A0M5J749 D3PG37 A0A2T7PKV1 A0A158NJM3 A0A1Z5L5C1 A0A194Q3K1 A0A0K2TX70 H9H6R1 A0A3M6V1N1 A0A1I8QDE5 A0A2A4IYD2 A0A1U7Q6J3 A0A3Q1JUX4 A0A2R5L585 A0A3L8DMA2 E2A971 B4MQJ7 A0A067QFM7 A0A061IBK7 A0A3B3YJ36 A0A087XCU0 A0A3B4T8X4 B1WAT5 A0A0S7LJQ2 A0A3B4XQ94 A0A0J7L311 U5EVW1 A0A3B4X0R8 E0VTU3 C3Y656 A0A067QYE5 K7J9H7 D2A3F2 A0A383Z5F2 A0A0A0V5H3 V5I9N6 A0A3B3Q3F5 Q6DDI3 A0A1L8GCX7 A0A383Z5B6 A0A2B4SAC1 A0A2A4K689 K1R7F9 A0A023EK84 W8ATQ8 A0A293MV59 A0A1Y1L5M1 M3ZCS7 T1E2B6 E2BYU7 A0A1W5B0K8 A0A3S2N5Y2 A0A1I8PKV3 A0A3Q1BMI7 A0A0L0CN39 A0A3P9N9K2 A0A3S1CB08 A0A076KUR5 A0A023EK90
Ontologies
GO
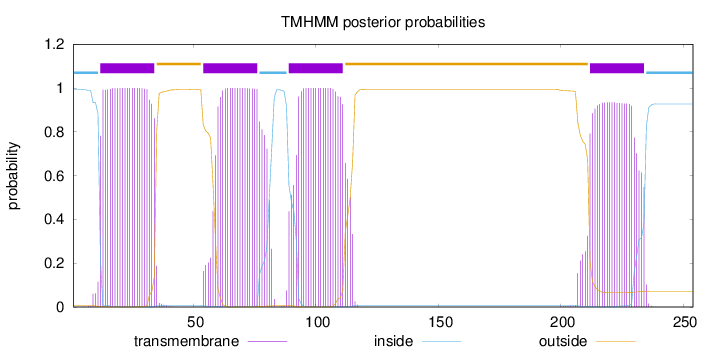
Topology
Subcellular location
Membrane
Length:
254
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
89.9819200000002
Exp number, first 60 AAs:
25.79312
Total prob of N-in:
0.99286
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 53
TMhelix
54 - 76
inside
77 - 88
TMhelix
89 - 111
outside
112 - 211
TMhelix
212 - 234
inside
235 - 254
Population Genetic Test Statistics
Pi
281.219599
Theta
203.036898
Tajima's D
1.202818
CLR
1.777908
CSRT
0.710264486775661
Interpretation
Uncertain
