Gene
KWMTBOMO14253
Pre Gene Modal
BGIBMGA002160
Annotation
PREDICTED:_putative_fatty_acyl-CoA_reductase_CG5065_isoform_X1_[Bombyx_mori]
Full name
Fatty acyl-CoA reductase
+ More
Putative fatty acyl-CoA reductase CG5065
Putative fatty acyl-CoA reductase CG5065
Location in the cell
Mitochondrial Reliability : 1.463 Nuclear Reliability : 1.073
Sequence
CDS
ATGGCTGATATGTCAGAAGAGGCTCCGATCCTTCCAAGGTTCTACGCGGGGAGATCGGTCTTCATTACGGGAGGCACCGGCTTTATGGGAAAGGTGTTGGTGGAGCGTCTACTGTCCACGTGCGATGATATCCAGGAGGTGTTCCTCCTCGTGCGGGAGAAGAAGGACGTGCCGCCAGAGAAGAGGCTCGCTCAGTTCAAGCAGTGTCAGTACACGAGGGTAATATGTGTAATATTCGAGAAGGTTCGGGAGCGGTGTCCGTGGCAGCTCGACAAGCTGCGCGTGCTCCCCGGGAACCTGGAGCGACCCCTGCTGGGACTCGAACCGGTGACTATCGCTCGACTACACCAGCCTACAGAGCGCGTCCGGTGCCCGCAGGTGTCGGTGGTGTTCCACAACGCTGCGACCCTCAAGTTCGACGAGGCTCTACGCAAGGCGGTCGACCAGAACGTGCGGGGCCTCCTGTCCCTCGTCGACGTGTGCGACGCCCTGCCGAACCTCGAGGCGCTGGTGCACGTGTCCACTGCGTACAGCAACGCGGAGCTGTCGAGGGTGGAGGAGCGCGTGTACGCGGCGCCGGTGCCGCTGCAGCAGCTGCTGGCGCTGGCCGACGCGCTGCCGGACCAGCTCGCCGCCGACCTCACGCCCAGGTACATCCACCCGAAGCCGAACACTTACACGTTCACGAAGGCGATGGCGGAGTGCGTGCTGCAGGAGAGGACCAGCCGCCGCTACCCGGTCGCCATCTTCCGACCGACCATAGTGATATCAGCAGACCGGCACCCGGTCCCGGGGTGGATAGAGAACTTGAACGGGCCGAGCGGCGTCGTCGTGGGGGTCGGCAAGGGGCTGATGCACGTGTTCAGAGCGAAGCTCGCGCTGCGGGCCGACTTCATCCCGGTCGACGTCGTCGTGGACAACATGATCGCCGTTGCCTGGGAGACGGCCTCAGAACCGTGCGGCTCCTTGCGCGTGTACAACTGCGCGTCGGGCGAGCACGCGACGCGCTTATCGGACTTCAGAGCCACCGCAGTCCGCGTGCTCCGGAGCCTCCCGCTGGACTCAGGACTCTCCTATCCCTTTCTGATCGTGGTGCAGAACAGGTTCGTGTACCGGCTGCTGGAGTTCGTGCTGCAGACGGCGCCGCTGCACGTGGCCGAGTGCTTGCGGCAGACCGACGGGCAGAAGGCGAGGTTGAGTTTCACAACGGCAGGGCGGCGCATACGAGCGATGACGGAGGTGCTGCACTTCTTCGTGCTGCGCGAGTGGACGTTCCCGTGCGACAACATCCGGCGGCTGCGGCGGCGGCTCACGGCGCGCGACCGCCGCATCTACCACCTGGACGTCGCGGAGGTCCATTGGGACGAATTATATTTAAACTTCGTGAAAGGAACAAGAAAATATTTACTTCAGGAAAAGGAAGAAGATTTGAAAGAAGCGCAAAAAAGAATGAAGAGATTGCGTCTCGTCCACTACACCACCATCGTGTTCCTCGTCTTACTATCCTACAAACTAATTAAGTTTTTCCTAAATTTGTTTATGAACAACTAA
Protein
MADMSEEAPILPRFYAGRSVFITGGTGFMGKVLVERLLSTCDDIQEVFLLVREKKDVPPEKRLAQFKQCQYTRVICVIFEKVRERCPWQLDKLRVLPGNLERPLLGLEPVTIARLHQPTERVRCPQVSVVFHNAATLKFDEALRKAVDQNVRGLLSLVDVCDALPNLEALVHVSTAYSNAELSRVEERVYAAPVPLQQLLALADALPDQLAADLTPRYIHPKPNTYTFTKAMAECVLQERTSRRYPVAIFRPTIVISADRHPVPGWIENLNGPSGVVVGVGKGLMHVFRAKLALRADFIPVDVVVDNMIAVAWETASEPCGSLRVYNCASGEHATRLSDFRATAVRVLRSLPLDSGLSYPFLIVVQNRFVYRLLEFVLQTAPLHVAECLRQTDGQKARLSFTTAGRRIRAMTEVLHFFVLREWTFPCDNIRRLRRRLTARDRRIYHLDVAEVHWDELYLNFVKGTRKYLLQEKEEDLKEAQKRMKRLRLVHYTTIVFLVLLSYKLIKFFLNLFMNN
Summary
Description
Catalyzes the reduction of fatty acyl-CoA to fatty alcohols.
Catalyzes the reduction of C16 or C18 fatty acyl-CoA to fatty alcohols.
Catalyzes the reduction of C16 or C18 fatty acyl-CoA to fatty alcohols.
Catalytic Activity
a long-chain fatty acyl-CoA + 2 H(+) + 2 NADPH = a long-chain primary fatty alcohol + CoA + 2 NADP(+)
Similarity
Belongs to the fatty acyl-CoA reductase family.
Keywords
Complete proteome
Lipid biosynthesis
Lipid metabolism
Membrane
NADP
Oxidoreductase
Peroxisome
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Putative fatty acyl-CoA reductase CG5065
Uniprot
H9IY27
A0A212FG68
A0A068FJB7
A0A0F6Q104
A0A2W1BU43
A0A0F6Q4L1
+ More
A0A2W1BD38 A0A0F6Q1K5 H9JNR0 A0A291P0Q0 D9D5H9 A0A386QW76 A0A194PTQ4 A0A0P0EA69 A0A2H1WNT2 A0A0P0EW34 A0A1V0M880 D7P5E4 U5KFU0 A0A3S2LSJ5 A0A2W1B0E2 A0A1L8D6B8 A0A291P0V6 A0A0F6Q1I7 A0A194RN00 A0A2A4JGI4 A0A3S2NBD7 A0A087TLH2 A0A023F1N4 E9GTL9 B4H764 Q28WN8 T1HPB8 A0A069DVI2 A0A1I8PNE0 A0A0N1IPU7 A0A3B0J1M1 B4KSN0 B4P5X9 B3NPG3 B3MDK3 B4LPA3 A0A1I8M522 T1PAL6 A0A0P5YBQ5 B4JWM8 A0A1W4UXP9 A0A0P5QJM8 A0A194PSL9 W8AXT4 A0A034VN48 A0A0K8U8F7 T1KB77 B4HT68 E0VSV6 A0A0J9RE92 E1JH83 A1ZAI5 A0A336LRW3 A0A0A1WIW0 A0A336MZZ9 B4MIU8 A0A212F418 A0A291P0J5 A0A1A9X2G9 A0A0M4EVU9 T1JSH5 A0A147BN16 A0A0P6ITJ7 A0A0L0BVZ6 A0A1B6EM21 A0A1B6G8A3 A0A1A9VQ82 A0A1B6GUZ4 A0A2J7PLY4 A0A0A9VYR3 A0A2L2YHA0 A0A1B6JB63 A0A1B0ARC3 A0A0A9VVQ7 A0A0C9QJE0 A0A131YLS0 A0A1B6KYT4 E9GMI7 A0A1J1IBH8 A0A0P0EM43 K7J1Z7 A0A069DV62 A7SBJ0 A0A3Q8FTW8 A0A1W4WEK2 A0A023GNB8 A0A023F1V0 A0A2J7PM67 E2B9J0 A0A132AJS7 E0VLJ7 A0A1E1X4I5 G3KIJ9
A0A2W1BD38 A0A0F6Q1K5 H9JNR0 A0A291P0Q0 D9D5H9 A0A386QW76 A0A194PTQ4 A0A0P0EA69 A0A2H1WNT2 A0A0P0EW34 A0A1V0M880 D7P5E4 U5KFU0 A0A3S2LSJ5 A0A2W1B0E2 A0A1L8D6B8 A0A291P0V6 A0A0F6Q1I7 A0A194RN00 A0A2A4JGI4 A0A3S2NBD7 A0A087TLH2 A0A023F1N4 E9GTL9 B4H764 Q28WN8 T1HPB8 A0A069DVI2 A0A1I8PNE0 A0A0N1IPU7 A0A3B0J1M1 B4KSN0 B4P5X9 B3NPG3 B3MDK3 B4LPA3 A0A1I8M522 T1PAL6 A0A0P5YBQ5 B4JWM8 A0A1W4UXP9 A0A0P5QJM8 A0A194PSL9 W8AXT4 A0A034VN48 A0A0K8U8F7 T1KB77 B4HT68 E0VSV6 A0A0J9RE92 E1JH83 A1ZAI5 A0A336LRW3 A0A0A1WIW0 A0A336MZZ9 B4MIU8 A0A212F418 A0A291P0J5 A0A1A9X2G9 A0A0M4EVU9 T1JSH5 A0A147BN16 A0A0P6ITJ7 A0A0L0BVZ6 A0A1B6EM21 A0A1B6G8A3 A0A1A9VQ82 A0A1B6GUZ4 A0A2J7PLY4 A0A0A9VYR3 A0A2L2YHA0 A0A1B6JB63 A0A1B0ARC3 A0A0A9VVQ7 A0A0C9QJE0 A0A131YLS0 A0A1B6KYT4 E9GMI7 A0A1J1IBH8 A0A0P0EM43 K7J1Z7 A0A069DV62 A7SBJ0 A0A3Q8FTW8 A0A1W4WEK2 A0A023GNB8 A0A023F1V0 A0A2J7PM67 E2B9J0 A0A132AJS7 E0VLJ7 A0A1E1X4I5 G3KIJ9
EC Number
1.2.1.84
Pubmed
19121390
22118469
26385554
28756777
28986331
26354079
+ More
26445454 28349297 20534481 24053512 25474469 21292972 17994087 15632085 26334808 17550304 18057021 25315136 24495485 25348373 20566863 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 29652888 26108605 25401762 26561354 26830274 20075255 17615350 20798317 26555130 28503490 21918929
26445454 28349297 20534481 24053512 25474469 21292972 17994087 15632085 26334808 17550304 18057021 25315136 24495485 25348373 20566863 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 29652888 26108605 25401762 26561354 26830274 20075255 17615350 20798317 26555130 28503490 21918929
EMBL
BABH01029849
AGBW02008707
OWR52728.1
KJ622059
AID66647.1
KP237929
+ More
AKD01782.1 KZ149962 PZC76300.1 KP237910 AKD01763.1 KZ150430 PZC70906.1 KP237936 MF687598 AKD01789.1 ATJ44524.1 BABH01035236 MF687540 ATJ44466.1 GU808255 ADI82779.1 MG585940 AYE54244.1 KQ459593 KPI96353.1 KT261705 ALJ30245.1 ODYU01009923 SOQ54667.1 KT261700 ALJ30240.1 KU755481 ARD71191.1 GQ907233 ADD62440.1 JX989157 AGR49316.1 RSAL01000287 RVE42944.1 KZ150513 PZC70632.1 GEYN01000138 JAV01991.1 MF687597 ATJ44523.1 KP237916 AKD01769.1 KQ460152 KPJ17376.1 NWSH01001610 PCG70694.1 RSAL01000280 RVE43065.1 KK115764 KFM65961.1 GBBI01003791 JAC14921.1 GL732564 EFX77189.1 CH479216 EDW33597.1 CM000071 EAL26629.1 ACPB03012985 GBGD01001172 JAC87717.1 KQ460036 KPJ18402.1 OUUW01000001 SPP72772.1 CH933808 EDW10529.2 CM000158 EDW91895.1 CH954179 EDV55730.1 CH902619 EDV37466.1 KPU76846.1 CH940648 EDW60212.2 KA645200 AFP59829.1 GDIP01061996 LRGB01000024 JAM41719.1 KZS21332.1 CH916375 EDV98366.1 GDIQ01123144 JAL28582.1 KPI96431.1 GAMC01012755 JAB93800.1 GAKP01015960 GAKP01015959 GAKP01015958 GAKP01015956 JAC42993.1 GDHF01029367 GDHF01008158 JAI22947.1 JAI44156.1 CAEY01001946 CH480816 EDW48169.1 DS235758 EEB16462.1 CM002911 KMY94302.1 AE013599 ACZ94440.1 AAF57974.1 UFQT01000095 SSX19701.1 GBXI01017222 GBXI01017048 GBXI01015288 GBXI01014990 GBXI01003875 JAC97069.1 JAC97243.1 JAC99003.1 JAC99301.1 JAD10417.1 UFQT01004920 SSX35816.1 CH963719 EDW72037.1 AGBW02010466 OWR48453.1 MF687541 ATJ44467.1 CP012524 ALC42075.1 CAEY01000461 GEGO01003258 JAR92146.1 GDIQ01005343 JAN89394.1 JRES01001254 KNC24212.1 GECZ01030782 JAS38987.1 GECZ01011099 JAS58670.1 GECZ01003528 JAS66241.1 NEVH01024421 PNF17348.1 GBHO01043288 GBRD01007683 JAG00316.1 JAG58138.1 IAAA01029277 IAAA01029278 LAA07536.1 GECU01011206 JAS96500.1 JXJN01002319 JXJN01002320 GBHO01043282 JAG00322.1 GBYB01014923 GBYB01014924 JAG84690.1 JAG84691.1 GEDV01009049 JAP79508.1 GEBQ01023412 JAT16565.1 GL732552 EFX79381.1 CVRI01000047 CRK97592.1 KT261695 ALJ30235.1 GBGD01001183 JAC87706.1 DS469616 EDO38930.1 MG573164 AWJ25026.1 GBBM01000775 JAC34643.1 GBBI01003721 JAC14991.1 PNF17417.1 GL446556 EFN87623.1 JXLN01016764 KPM11252.1 DS235271 EEB14253.1 GFAC01005008 JAT94180.1 JN243756 AEO89346.1
AKD01782.1 KZ149962 PZC76300.1 KP237910 AKD01763.1 KZ150430 PZC70906.1 KP237936 MF687598 AKD01789.1 ATJ44524.1 BABH01035236 MF687540 ATJ44466.1 GU808255 ADI82779.1 MG585940 AYE54244.1 KQ459593 KPI96353.1 KT261705 ALJ30245.1 ODYU01009923 SOQ54667.1 KT261700 ALJ30240.1 KU755481 ARD71191.1 GQ907233 ADD62440.1 JX989157 AGR49316.1 RSAL01000287 RVE42944.1 KZ150513 PZC70632.1 GEYN01000138 JAV01991.1 MF687597 ATJ44523.1 KP237916 AKD01769.1 KQ460152 KPJ17376.1 NWSH01001610 PCG70694.1 RSAL01000280 RVE43065.1 KK115764 KFM65961.1 GBBI01003791 JAC14921.1 GL732564 EFX77189.1 CH479216 EDW33597.1 CM000071 EAL26629.1 ACPB03012985 GBGD01001172 JAC87717.1 KQ460036 KPJ18402.1 OUUW01000001 SPP72772.1 CH933808 EDW10529.2 CM000158 EDW91895.1 CH954179 EDV55730.1 CH902619 EDV37466.1 KPU76846.1 CH940648 EDW60212.2 KA645200 AFP59829.1 GDIP01061996 LRGB01000024 JAM41719.1 KZS21332.1 CH916375 EDV98366.1 GDIQ01123144 JAL28582.1 KPI96431.1 GAMC01012755 JAB93800.1 GAKP01015960 GAKP01015959 GAKP01015958 GAKP01015956 JAC42993.1 GDHF01029367 GDHF01008158 JAI22947.1 JAI44156.1 CAEY01001946 CH480816 EDW48169.1 DS235758 EEB16462.1 CM002911 KMY94302.1 AE013599 ACZ94440.1 AAF57974.1 UFQT01000095 SSX19701.1 GBXI01017222 GBXI01017048 GBXI01015288 GBXI01014990 GBXI01003875 JAC97069.1 JAC97243.1 JAC99003.1 JAC99301.1 JAD10417.1 UFQT01004920 SSX35816.1 CH963719 EDW72037.1 AGBW02010466 OWR48453.1 MF687541 ATJ44467.1 CP012524 ALC42075.1 CAEY01000461 GEGO01003258 JAR92146.1 GDIQ01005343 JAN89394.1 JRES01001254 KNC24212.1 GECZ01030782 JAS38987.1 GECZ01011099 JAS58670.1 GECZ01003528 JAS66241.1 NEVH01024421 PNF17348.1 GBHO01043288 GBRD01007683 JAG00316.1 JAG58138.1 IAAA01029277 IAAA01029278 LAA07536.1 GECU01011206 JAS96500.1 JXJN01002319 JXJN01002320 GBHO01043282 JAG00322.1 GBYB01014923 GBYB01014924 JAG84690.1 JAG84691.1 GEDV01009049 JAP79508.1 GEBQ01023412 JAT16565.1 GL732552 EFX79381.1 CVRI01000047 CRK97592.1 KT261695 ALJ30235.1 GBGD01001183 JAC87706.1 DS469616 EDO38930.1 MG573164 AWJ25026.1 GBBM01000775 JAC34643.1 GBBI01003721 JAC14991.1 PNF17417.1 GL446556 EFN87623.1 JXLN01016764 KPM11252.1 DS235271 EEB14253.1 GFAC01005008 JAT94180.1 JN243756 AEO89346.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000283053
UP000053240
UP000218220
+ More
UP000054359 UP000000305 UP000008744 UP000001819 UP000015103 UP000095300 UP000268350 UP000009192 UP000002282 UP000008711 UP000007801 UP000008792 UP000095301 UP000076858 UP000001070 UP000192221 UP000015104 UP000001292 UP000009046 UP000000803 UP000007798 UP000091820 UP000092553 UP000037069 UP000078200 UP000235965 UP000092460 UP000183832 UP000002358 UP000001593 UP000192223 UP000008237
UP000054359 UP000000305 UP000008744 UP000001819 UP000015103 UP000095300 UP000268350 UP000009192 UP000002282 UP000008711 UP000007801 UP000008792 UP000095301 UP000076858 UP000001070 UP000192221 UP000015104 UP000001292 UP000009046 UP000000803 UP000007798 UP000091820 UP000092553 UP000037069 UP000078200 UP000235965 UP000092460 UP000183832 UP000002358 UP000001593 UP000192223 UP000008237
Interpro
SUPFAM
SSF51735
SSF51735
CDD
ProteinModelPortal
H9IY27
A0A212FG68
A0A068FJB7
A0A0F6Q104
A0A2W1BU43
A0A0F6Q4L1
+ More
A0A2W1BD38 A0A0F6Q1K5 H9JNR0 A0A291P0Q0 D9D5H9 A0A386QW76 A0A194PTQ4 A0A0P0EA69 A0A2H1WNT2 A0A0P0EW34 A0A1V0M880 D7P5E4 U5KFU0 A0A3S2LSJ5 A0A2W1B0E2 A0A1L8D6B8 A0A291P0V6 A0A0F6Q1I7 A0A194RN00 A0A2A4JGI4 A0A3S2NBD7 A0A087TLH2 A0A023F1N4 E9GTL9 B4H764 Q28WN8 T1HPB8 A0A069DVI2 A0A1I8PNE0 A0A0N1IPU7 A0A3B0J1M1 B4KSN0 B4P5X9 B3NPG3 B3MDK3 B4LPA3 A0A1I8M522 T1PAL6 A0A0P5YBQ5 B4JWM8 A0A1W4UXP9 A0A0P5QJM8 A0A194PSL9 W8AXT4 A0A034VN48 A0A0K8U8F7 T1KB77 B4HT68 E0VSV6 A0A0J9RE92 E1JH83 A1ZAI5 A0A336LRW3 A0A0A1WIW0 A0A336MZZ9 B4MIU8 A0A212F418 A0A291P0J5 A0A1A9X2G9 A0A0M4EVU9 T1JSH5 A0A147BN16 A0A0P6ITJ7 A0A0L0BVZ6 A0A1B6EM21 A0A1B6G8A3 A0A1A9VQ82 A0A1B6GUZ4 A0A2J7PLY4 A0A0A9VYR3 A0A2L2YHA0 A0A1B6JB63 A0A1B0ARC3 A0A0A9VVQ7 A0A0C9QJE0 A0A131YLS0 A0A1B6KYT4 E9GMI7 A0A1J1IBH8 A0A0P0EM43 K7J1Z7 A0A069DV62 A7SBJ0 A0A3Q8FTW8 A0A1W4WEK2 A0A023GNB8 A0A023F1V0 A0A2J7PM67 E2B9J0 A0A132AJS7 E0VLJ7 A0A1E1X4I5 G3KIJ9
A0A2W1BD38 A0A0F6Q1K5 H9JNR0 A0A291P0Q0 D9D5H9 A0A386QW76 A0A194PTQ4 A0A0P0EA69 A0A2H1WNT2 A0A0P0EW34 A0A1V0M880 D7P5E4 U5KFU0 A0A3S2LSJ5 A0A2W1B0E2 A0A1L8D6B8 A0A291P0V6 A0A0F6Q1I7 A0A194RN00 A0A2A4JGI4 A0A3S2NBD7 A0A087TLH2 A0A023F1N4 E9GTL9 B4H764 Q28WN8 T1HPB8 A0A069DVI2 A0A1I8PNE0 A0A0N1IPU7 A0A3B0J1M1 B4KSN0 B4P5X9 B3NPG3 B3MDK3 B4LPA3 A0A1I8M522 T1PAL6 A0A0P5YBQ5 B4JWM8 A0A1W4UXP9 A0A0P5QJM8 A0A194PSL9 W8AXT4 A0A034VN48 A0A0K8U8F7 T1KB77 B4HT68 E0VSV6 A0A0J9RE92 E1JH83 A1ZAI5 A0A336LRW3 A0A0A1WIW0 A0A336MZZ9 B4MIU8 A0A212F418 A0A291P0J5 A0A1A9X2G9 A0A0M4EVU9 T1JSH5 A0A147BN16 A0A0P6ITJ7 A0A0L0BVZ6 A0A1B6EM21 A0A1B6G8A3 A0A1A9VQ82 A0A1B6GUZ4 A0A2J7PLY4 A0A0A9VYR3 A0A2L2YHA0 A0A1B6JB63 A0A1B0ARC3 A0A0A9VVQ7 A0A0C9QJE0 A0A131YLS0 A0A1B6KYT4 E9GMI7 A0A1J1IBH8 A0A0P0EM43 K7J1Z7 A0A069DV62 A7SBJ0 A0A3Q8FTW8 A0A1W4WEK2 A0A023GNB8 A0A023F1V0 A0A2J7PM67 E2B9J0 A0A132AJS7 E0VLJ7 A0A1E1X4I5 G3KIJ9
PDB
4U5Q
E-value=0.00250501,
Score=98
Ontologies
GO
PANTHER
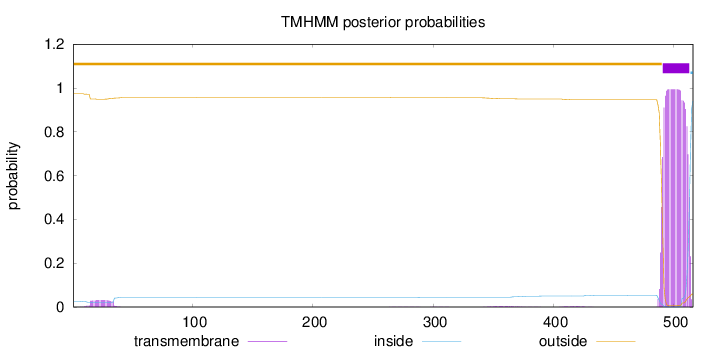
Topology
Subcellular location
Peroxisome membrane
Length:
516
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.1481
Exp number, first 60 AAs:
0.64282
Total prob of N-in:
0.02616
outside
1 - 490
TMhelix
491 - 513
inside
514 - 516
Population Genetic Test Statistics
Pi
12.926046
Theta
13.248354
Tajima's D
-0.272085
CLR
0.896463
CSRT
0.296185190740463
Interpretation
Uncertain
