Pre Gene Modal
BGIBMGA011166
Annotation
protein_kinase_C_inhibitor_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.875
Sequence
CDS
ATGTTCAACAAAGTTTTCCAACGCGCACTAAAATTTCGATTGAAAAAGACTTATCCTACGAACATCTATGATCTGTCTCGGGGTTCTGCTGTGGCTGTGAAGCGGCCGTACAGCGACGAAGTCCGCAGAGCACACGAAACGACCACTACGATCGGGCCCACAATATTCGACAAGATCATATCTAAGGAGATCAGAGCCGATATCATATACGAAGATGATTTATGTCTTGCTTTTAACGACATAGCACCGCAGGCGCCAGTACATTTTCTTGTTATTCCAAAGCGTAGAATAGCAAGGTTGCAGGATGCGGAGAACAACGACAACGAGCTGCTAGGTCACTTGATGCTGGTAGCCCGGTCACTCGGTGCCCAAAGAGCTCCTTCAGGATGGCGTCTTGTGGTTAACAATGGCAAAGACGGAGCTCAAAGTGTTTATCATCTACACCTTCATGTTCTGGGTGGAAGACAGATGGGCTGGCCACCTGGAATTTCTGGTTCTAAAGAACTAGATTCACAATAG
Protein
MFNKVFQRALKFRLKKTYPTNIYDLSRGSAVAVKRPYSDEVRRAHETTTTIGPTIFDKIISKEIRADIIYEDDLCLAFNDIAPQAPVHFLVIPKRRIARLQDAENNDNELLGHLMLVARSLGAQRAPSGWRLVVNNGKDGAQSVYHLHLHVLGGRQMGWPPGISGSKELDSQ
Summary
Uniprot
Q2F5W9
A0A2A4JF27
A0A2W1B8Y9
A0A0L7KRN1
A0A3S2N6U4
A0A212FG76
+ More
S4P7I2 A0A1L8DL87 A0A1B0D3L1 A0A194RII9 A0A1B0CHZ6 E0VVC2 A0A183B396 A0A3S5AFK7 A0A183PNH8 A0A183JZT4 A0A3Q0KFK0 A0A3M7SEU5 D6W8N6 A0A3R7MYQ8 A0A2M4C2H8 A0A2M4C2E6 A0A3Q3LIC1 A0A3Q3GWT5 A0A3Q0TGJ7 A0A3B4GMR3 A0A3Q2WIS6 A0A3P8QKN0 A0A3P9C820 C1LLG3 A0A2M4C3B5 C1L7D7 A0A3B3TPW3 A0A3Q2D3Z7 A0A3Q3J2B6 A0A3Q1CTP9 A0A3P9C7I4 A0A3Q2STK4 A0A3B4VJN4 A0A3P9NQH9 C3ZJE2 A0A0F8ASD9 A0A3B3XJ85 A0A3B4GMD3 A0A131XLS3 A0A3Q4I169 A0A3Q2D4B3 A0A3B4WYJ4 A0A3P9KIW8 Q642J0 A0A3B5KFQ1 A0A0X3PEB7 A0A315VAA8 Q5DHW9 A0A2H1C3L0 A0A146VCP1 A0A3Q1CSZ1 A0A1A8QCI6 A0A1A7Y0R3 A0A3P9KIW1 A0A3B3V8F2 A0A3B5AKA1 A0A087XMV7 A0A3Q4IER6 A0A1A8V4Q4 A0A3B3C2J2 A0A3B3QPM9 A0A2I4CI71 A0A3P9M0V9 A0A3P8YD88 A0A1A8NVR5 A0A1A8L112
S4P7I2 A0A1L8DL87 A0A1B0D3L1 A0A194RII9 A0A1B0CHZ6 E0VVC2 A0A183B396 A0A3S5AFK7 A0A183PNH8 A0A183JZT4 A0A3Q0KFK0 A0A3M7SEU5 D6W8N6 A0A3R7MYQ8 A0A2M4C2H8 A0A2M4C2E6 A0A3Q3LIC1 A0A3Q3GWT5 A0A3Q0TGJ7 A0A3B4GMR3 A0A3Q2WIS6 A0A3P8QKN0 A0A3P9C820 C1LLG3 A0A2M4C3B5 C1L7D7 A0A3B3TPW3 A0A3Q2D3Z7 A0A3Q3J2B6 A0A3Q1CTP9 A0A3P9C7I4 A0A3Q2STK4 A0A3B4VJN4 A0A3P9NQH9 C3ZJE2 A0A0F8ASD9 A0A3B3XJ85 A0A3B4GMD3 A0A131XLS3 A0A3Q4I169 A0A3Q2D4B3 A0A3B4WYJ4 A0A3P9KIW8 Q642J0 A0A3B5KFQ1 A0A0X3PEB7 A0A315VAA8 Q5DHW9 A0A2H1C3L0 A0A146VCP1 A0A3Q1CSZ1 A0A1A8QCI6 A0A1A7Y0R3 A0A3P9KIW1 A0A3B3V8F2 A0A3B5AKA1 A0A087XMV7 A0A3Q4IER6 A0A1A8V4Q4 A0A3B3C2J2 A0A3B3QPM9 A0A2I4CI71 A0A3P9M0V9 A0A3P8YD88 A0A1A8NVR5 A0A1A8L112
Pubmed
EMBL
BABH01035236
DQ311304
ABD36248.1
NWSH01001610
PCG70695.1
KZ150430
+ More
PZC70905.1 JTDY01006774 KOB65701.1 RSAL01000287 RVE42943.1 AGBW02008707 OWR52727.1 GAIX01004594 JAA87966.1 GFDF01006903 JAV07181.1 AJVK01002952 KQ460152 KPJ17377.1 AJWK01012924 DS235809 EEB17328.1 UZAN01055679 VDP90953.1 CAAALY010091812 VEL28026.1 UZAL01036478 VDP69897.1 UZAK01032657 VDP29831.1 REGN01001501 RNA34286.1 KQ971312 EEZ98381.1 QCYY01002091 ROT72903.1 GGFJ01010353 MBW59494.1 GGFJ01010355 MBW59496.1 FN314882 FN319815 FN319816 FN319817 CAX75541.1 GGFJ01010357 MBW59498.1 FN314883 CAX70615.1 GG666632 EEN47284.1 KQ042852 KKF09501.1 GEFH01001493 JAP67088.1 BX537303 BC081526 BC164932 AAH81526.1 AAI64932.1 GEEE01013177 JAP50048.1 NHOQ01002573 PWA15900.1 AY812855 AAW24587.1 KZ431762 PIS81302.1 GCES01071464 JAR14859.1 HAEI01004979 SBR91231.1 HADW01016455 HADX01001442 SBP23674.1 AYCK01007159 AYCK01007160 HAEJ01013761 SBS54218.1 HAEH01004130 SBR73108.1 HAEF01000761 HAEG01002857 SBR38143.1
PZC70905.1 JTDY01006774 KOB65701.1 RSAL01000287 RVE42943.1 AGBW02008707 OWR52727.1 GAIX01004594 JAA87966.1 GFDF01006903 JAV07181.1 AJVK01002952 KQ460152 KPJ17377.1 AJWK01012924 DS235809 EEB17328.1 UZAN01055679 VDP90953.1 CAAALY010091812 VEL28026.1 UZAL01036478 VDP69897.1 UZAK01032657 VDP29831.1 REGN01001501 RNA34286.1 KQ971312 EEZ98381.1 QCYY01002091 ROT72903.1 GGFJ01010353 MBW59494.1 GGFJ01010355 MBW59496.1 FN314882 FN319815 FN319816 FN319817 CAX75541.1 GGFJ01010357 MBW59498.1 FN314883 CAX70615.1 GG666632 EEN47284.1 KQ042852 KKF09501.1 GEFH01001493 JAP67088.1 BX537303 BC081526 BC164932 AAH81526.1 AAI64932.1 GEEE01013177 JAP50048.1 NHOQ01002573 PWA15900.1 AY812855 AAW24587.1 KZ431762 PIS81302.1 GCES01071464 JAR14859.1 HAEI01004979 SBR91231.1 HADW01016455 HADX01001442 SBP23674.1 AYCK01007159 AYCK01007160 HAEJ01013761 SBS54218.1 HAEH01004130 SBR73108.1 HAEF01000761 HAEG01002857 SBR38143.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000007151
UP000092462
+ More
UP000053240 UP000092461 UP000009046 UP000050740 UP000050791 UP000050789 UP000008854 UP000276133 UP000007266 UP000283509 UP000261640 UP000261660 UP000261340 UP000261460 UP000264840 UP000265100 UP000265160 UP000261500 UP000265020 UP000261600 UP000257160 UP000265000 UP000261420 UP000242638 UP000001554 UP000261480 UP000261580 UP000261360 UP000265180 UP000000437 UP000005226 UP000261400 UP000028760 UP000261560 UP000261540 UP000192220 UP000265140
UP000053240 UP000092461 UP000009046 UP000050740 UP000050791 UP000050789 UP000008854 UP000276133 UP000007266 UP000283509 UP000261640 UP000261660 UP000261340 UP000261460 UP000264840 UP000265100 UP000265160 UP000261500 UP000265020 UP000261600 UP000257160 UP000265000 UP000261420 UP000242638 UP000001554 UP000261480 UP000261580 UP000261360 UP000265180 UP000000437 UP000005226 UP000261400 UP000028760 UP000261560 UP000261540 UP000192220 UP000265140
PRIDE
Pfam
PF01230 HIT
Interpro
SUPFAM
SSF54197
SSF54197
Gene 3D
ProteinModelPortal
Q2F5W9
A0A2A4JF27
A0A2W1B8Y9
A0A0L7KRN1
A0A3S2N6U4
A0A212FG76
+ More
S4P7I2 A0A1L8DL87 A0A1B0D3L1 A0A194RII9 A0A1B0CHZ6 E0VVC2 A0A183B396 A0A3S5AFK7 A0A183PNH8 A0A183JZT4 A0A3Q0KFK0 A0A3M7SEU5 D6W8N6 A0A3R7MYQ8 A0A2M4C2H8 A0A2M4C2E6 A0A3Q3LIC1 A0A3Q3GWT5 A0A3Q0TGJ7 A0A3B4GMR3 A0A3Q2WIS6 A0A3P8QKN0 A0A3P9C820 C1LLG3 A0A2M4C3B5 C1L7D7 A0A3B3TPW3 A0A3Q2D3Z7 A0A3Q3J2B6 A0A3Q1CTP9 A0A3P9C7I4 A0A3Q2STK4 A0A3B4VJN4 A0A3P9NQH9 C3ZJE2 A0A0F8ASD9 A0A3B3XJ85 A0A3B4GMD3 A0A131XLS3 A0A3Q4I169 A0A3Q2D4B3 A0A3B4WYJ4 A0A3P9KIW8 Q642J0 A0A3B5KFQ1 A0A0X3PEB7 A0A315VAA8 Q5DHW9 A0A2H1C3L0 A0A146VCP1 A0A3Q1CSZ1 A0A1A8QCI6 A0A1A7Y0R3 A0A3P9KIW1 A0A3B3V8F2 A0A3B5AKA1 A0A087XMV7 A0A3Q4IER6 A0A1A8V4Q4 A0A3B3C2J2 A0A3B3QPM9 A0A2I4CI71 A0A3P9M0V9 A0A3P8YD88 A0A1A8NVR5 A0A1A8L112
S4P7I2 A0A1L8DL87 A0A1B0D3L1 A0A194RII9 A0A1B0CHZ6 E0VVC2 A0A183B396 A0A3S5AFK7 A0A183PNH8 A0A183JZT4 A0A3Q0KFK0 A0A3M7SEU5 D6W8N6 A0A3R7MYQ8 A0A2M4C2H8 A0A2M4C2E6 A0A3Q3LIC1 A0A3Q3GWT5 A0A3Q0TGJ7 A0A3B4GMR3 A0A3Q2WIS6 A0A3P8QKN0 A0A3P9C820 C1LLG3 A0A2M4C3B5 C1L7D7 A0A3B3TPW3 A0A3Q2D3Z7 A0A3Q3J2B6 A0A3Q1CTP9 A0A3P9C7I4 A0A3Q2STK4 A0A3B4VJN4 A0A3P9NQH9 C3ZJE2 A0A0F8ASD9 A0A3B3XJ85 A0A3B4GMD3 A0A131XLS3 A0A3Q4I169 A0A3Q2D4B3 A0A3B4WYJ4 A0A3P9KIW8 Q642J0 A0A3B5KFQ1 A0A0X3PEB7 A0A315VAA8 Q5DHW9 A0A2H1C3L0 A0A146VCP1 A0A3Q1CSZ1 A0A1A8QCI6 A0A1A7Y0R3 A0A3P9KIW1 A0A3B3V8F2 A0A3B5AKA1 A0A087XMV7 A0A3Q4IER6 A0A1A8V4Q4 A0A3B3C2J2 A0A3B3QPM9 A0A2I4CI71 A0A3P9M0V9 A0A3P8YD88 A0A1A8NVR5 A0A1A8L112
PDB
3QGZ
E-value=1.78927e-38,
Score=395
Ontologies
GO
Topology
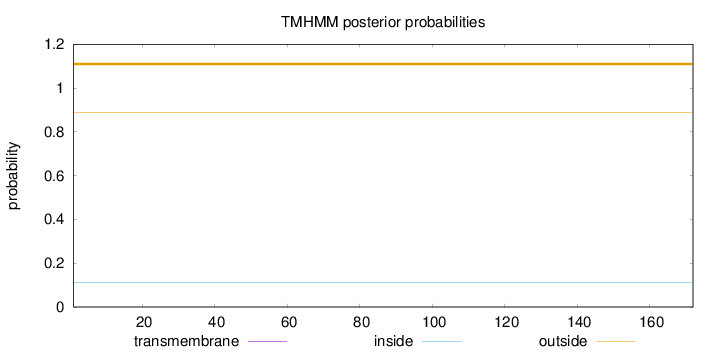
Length:
172
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00264
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11059
outside
1 - 172
Population Genetic Test Statistics
Pi
6.60968
Theta
12.508384
Tajima's D
-0.779651
CLR
0.190938
CSRT
0.184440777961102
Interpretation
Uncertain
