Gene
KWMTBOMO14243
Pre Gene Modal
BGIBMGA011102
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_son_of_sevenless_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.825
Sequence
CDS
ATGACCTCGCACGTCGAATGTGGTACATGTGTATGGGTGAGGTCGCTCGACGAGCGGGGGGGATCAGTGCTGGGAGTGCCTAAAGCACCGAGAGGGAACCGGGAGGGTCCGATAGGTTTCGGAGGTGGTGCGCGGTTGGACATCGCCTGCCCCCTTACAGTAATACCTAAAGGTTTCCTCTTGTCTCTTGCGCGATATAAAAACATCTATATTTTCAAGGTAAAACCATCCGAGCTGGTCGGCGCCGTGTGGACGAAAAAAGACAAGGAGAAGACTAGCCCTAACCTGTTGAAGATTATAAAGCATACGACCAACTTCACGCGCTGGATGGAGAAGTGGATAGTGGAGAGCGAGAACCTGGAGGAGCGCGTGGCGGTGGTGTGCCGCGTGCTGGAGCTGGGCGTGGCGCTGCTGCACCTCAACAACTTCAACGGCGTGCTGGCCGTCGTAGCCGCCGCCGGCAGCGCCTCCGTGCACAGACTCCGGGCCACCTTCCAGTTGATTCCTCTCCGCCTGCAAAAATACCTCGAGGAGCTCCGGGAGCTGAACACAGATCACTTCCGGAGATACCAGGAGAGGCTGAGGAGTATCAACCCTCCGTGCGTCCCCTTCTTCGGCATGTACCTCACCAACATCCTGCACATCGAGGAGGGGAACCTGGACTACCTTCCAAATTCCGAATTAATAAATTTCTCGAAGCGAAGAAAAGTGGCGGAGATCACGGGCGAGATCCAGCAGTACCAGAACCAGCCCTACTGCCTCACCGTGGAGCCCAGGACCAGGGCATTCCTGGAGTCGCTGGACCCGTTTCCCGGGAAAGACGACATCGAGATCACAAATTATCTGTACAACAAAAGTCTGGAAATAGAGCCCCGGCCGCCGGTCAAACAGATGCCGAAGTATCCCAGGAAGTACCCGGACCTGTCGCTGAAGCAGGTGAAGGCCAGCCGGAGGAACCAGCACGACCACTCCACCTGCGACGACAACCTCAGCGTGTCGCAGCTGTCCCCCACGGGCAGCTCGTGGGACGGCGCGTCGGTGGCGTCGCTGCCCATGCACCACGACGACCACAGAGGCGGCAGGGCGGAGGCCGGCAGCCCCAAGGCCGACGCGCGCTCGCTCAGCCAGCTCAGCCTCTTCGACAAGAAGCTCAGTCTGTTCGACAAGGGCAGGAGCCACAGGAACAGCGAGCCGCCCTCCCCCAGGGACAAGCACTCCCCCAGGCACGACCGGCCGGCGCCCGTCGAGAGAAAACCACAGAACTTATCGCCGCGGAACCCCATCGTCGACGCCGACTCCTTCTACACGAGCCGACACAGCCTCGAGCTCGGCGCCAGGAAGCACGAGAGTAAAGAGGAGAGCGTGCCCCCCCTTCTGCCGCGACACAGCGAGCCACCCCCAGCCCCGCCGCGCCGGATAGACACCCACCATGCGGCGCCGGGCGAGCAGCCGCCGGAGCCCCCGGACCGGCCGCAGCCCAGCCCCCAGCACTGCGAGCTCCTCAGGTGA
Protein
MTSHVECGTCVWVRSLDERGGSVLGVPKAPRGNREGPIGFGGGARLDIACPLTVIPKGFLLSLARYKNIYIFKVKPSELVGAVWTKKDKEKTSPNLLKIIKHTTNFTRWMEKWIVESENLEERVAVVCRVLELGVALLHLNNFNGVLAVVAAAGSASVHRLRATFQLIPLRLQKYLEELRELNTDHFRRYQERLRSINPPCVPFFGMYLTNILHIEEGNLDYLPNSELINFSKRRKVAEITGEIQQYQNQPYCLTVEPRTRAFLESLDPFPGKDDIEITNYLYNKSLEIEPRPPVKQMPKYPRKYPDLSLKQVKASRRNQHDHSTCDDNLSVSQLSPTGSSWDGASVASLPMHHDDHRGGRAEAGSPKADARSLSQLSLFDKKLSLFDKGRSHRNSEPPSPRDKHSPRHDRPAPVERKPQNLSPRNPIVDADSFYTSRHSLELGARKHESKEESVPPLLPRHSEPPPAPPRRIDTHHAAPGEQPPEPPDRPQPSPQHCELLR
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
IPR019804
Ras_G-nucl-exch_fac_CS
+ More
IPR011993 PH-like_dom_sf
IPR000651 Ras-like_Gua-exchang_fac_N
IPR007125 Histone_H2A/H2B/H3
IPR000219 DH-domain
IPR009072 Histone-fold
IPR036964 RASGEF_cat_dom_sf
IPR008937 Ras-like_GEF
IPR023578 Ras_GEF_dom_sf
IPR035899 DBL_dom_sf
IPR001895 RASGEF_cat_dom
IPR001849 PH_domain
IPR002119 Histone_H2A
IPR011993 PH-like_dom_sf
IPR000651 Ras-like_Gua-exchang_fac_N
IPR007125 Histone_H2A/H2B/H3
IPR000219 DH-domain
IPR009072 Histone-fold
IPR036964 RASGEF_cat_dom_sf
IPR008937 Ras-like_GEF
IPR023578 Ras_GEF_dom_sf
IPR035899 DBL_dom_sf
IPR001895 RASGEF_cat_dom
IPR001849 PH_domain
IPR002119 Histone_H2A
Gene 3D
ProteinModelPortal
PDB
1XDV
E-value=1.53712e-74,
Score=712
Ontologies
GO
PANTHER
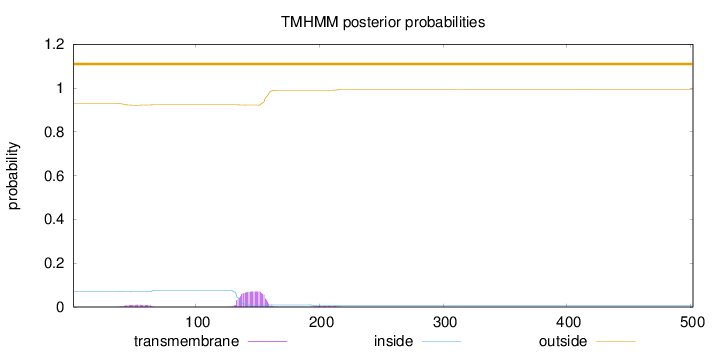
Topology
Length:
502
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.85286
Exp number, first 60 AAs:
0.16809
Total prob of N-in:
0.07134
outside
1 - 502
Population Genetic Test Statistics
Pi
6.383497
Theta
7.697207
Tajima's D
0.114782
CLR
0.560118
CSRT
0.420378981050947
Interpretation
Uncertain
