Gene
KWMTBOMO14237 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011108
Annotation
PREDICTED:_dehydrogenase/reductase_SDR_family_member_4_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.055
Sequence
CDS
ATGTCAACTTCAGCAAACCCTGTTCGTGCAATTCAGAGCACTCGGTTCAGTGGGAAGGTGGCTATTGTAACAGCATCGACTGAAGGAATTGGGTACGCAATAGCTAAGAGGCTGGGAAGTGAAGGTGCTTCCGTAGTCATCTGCAGCCGGAAAGAGAGCAACGTTGGGAAAGCTGTGCAGAGCCTCCGAAGCGAGGGCATTACAGTGGAAGGAGTCGTCTGTCACGTCGCCAATCATGAACATAGGAGACGATTATTTGAAGTGGCCACAAGTAAGTTTGGCGGAGTGGATATCCTGGTATCAAACGCCGCGGTCAATCCAGCGGTGGCTTCGATCTTAGATACCGACGAACAAGTCTGGGACAAAATATTTGACGTTAACGTGAAAAGCTCCTGGCTGTTGGCCAAGGAGGCGTATCCGGAGCTGGTCAAGAGAGGAGGGGGCAGTATCGTATTCATATCTTCGATTGCAGCGTATCAACCAACGGAGCCTCTAGGTGCCTATGGTGTTAGTAAAACGACGCTCCTTGGACTTACGAAGGCGATAGCCAACGAAACAGTACACGAGAACATCAGAGTGAACTGCGTCGCTCCCGGGATCGTCGTCACAAAGTTTGCATCCGCGATAACAAACTCCGAGGCTGGGGCGGAGAAGAGCCTATCGAATGTCCCATTGAAAAGATTCGGGAGACCAGAAGAAATTGCTGGTGCCGTAGCGTTTTTGGCTTCGGATGACGCCAGCTACATCACCGGAGAGACTGTTGTGGTTGCTGGAGGGGCGCCCGCACGACTTTGA
Protein
MSTSANPVRAIQSTRFSGKVAIVTASTEGIGYAIAKRLGSEGASVVICSRKESNVGKAVQSLRSEGITVEGVVCHVANHEHRRRLFEVATSKFGGVDILVSNAAVNPAVASILDTDEQVWDKIFDVNVKSSWLLAKEAYPELVKRGGGSIVFISSIAAYQPTEPLGAYGVSKTTLLGLTKAIANETVHENIRVNCVAPGIVVTKFASAITNSEAGAEKSLSNVPLKRFGRPEEIAGAVAFLASDDASYITGETVVVAGGAPARL
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JNK4
A0A2A4JK89
A0A194PWI4
D2SNW0
A0A194QUE6
A0A2H1VNY5
+ More
H9JQH9 A0A3S2L990 A0A212EM40 R4WDM8 A0A182X3R6 A0A182I3A3 A0A182XYV9 U5EWL2 A0A182VMY9 A0A182WJ87 A0A182KRB2 A0A182RSN1 Q7QJE4 A0A1Q3FJU4 A0A182MEK3 A0A084WN98 A0A1S4FCV9 A0A182NHV8 A0A182PQF5 A0A182IN68 Q176Z2 A0A0T6AUL9 A0A182GXS7 A0A182G5T3 A0A1B6C7C6 B0WXE6 D6WFA3 A0A182QY63 A0A182JV87 A0A1B6F9X4 A0A023EL45 W5JPB4 A0A1J1I894 A0A2M3ZBF2 A0A1S4ECP8 A0A2M4BW22 A0A1B6L3X8 A0A182FF41 A0A023FA77 A0A2M4BW41 A0A182UHE1 A0A0L0BKR1 D6WFA2 Q5FVX2 A0A026W8F7 A0A336LXI2 A0A2J7QXC6 A0A1L8ECQ8 A0A2M3ZJG7 A0A1L8EBM6 Q6DJN9 F7A1E4 A0A2G8K236 A0A0P4VRY8 A0A1L8HYS5 A0A1L8ECT5 A0A1I8NQW8 A0A0V0G5D1 A0A0N8DC99 W8C3I0 A0A0N8BJS9 A0A0P6I897 A0A067QR58 E9FVF3 A0A1L8EA07 A0A182SN43 A0A0P5T9G6 A0A087UTH3 A0A1I8N4U5 A0A0P5JEB6 A0A1A9WBP1 A0A0N8CAM7 A0A0P5UCX3 A0A2P6K321 A0A2C9JMD9 A0A1A9VN14 A0A0P5ULN5 A0A0K8SGQ3 A0A0M3QVW0 A0A1A9ZMI0 D3TNI9 A0A3S1BUX9 B4IX02 A0A2S2NMB6 A0A0A9Z575 A0A0N8EGT2 A0A0P6BMU8 A0A0A1XNH2 B4KWZ6 A0A2B4S7B5 A0A0K8WF41 B4HUD8 T1KNI7 B4LFQ3
H9JQH9 A0A3S2L990 A0A212EM40 R4WDM8 A0A182X3R6 A0A182I3A3 A0A182XYV9 U5EWL2 A0A182VMY9 A0A182WJ87 A0A182KRB2 A0A182RSN1 Q7QJE4 A0A1Q3FJU4 A0A182MEK3 A0A084WN98 A0A1S4FCV9 A0A182NHV8 A0A182PQF5 A0A182IN68 Q176Z2 A0A0T6AUL9 A0A182GXS7 A0A182G5T3 A0A1B6C7C6 B0WXE6 D6WFA3 A0A182QY63 A0A182JV87 A0A1B6F9X4 A0A023EL45 W5JPB4 A0A1J1I894 A0A2M3ZBF2 A0A1S4ECP8 A0A2M4BW22 A0A1B6L3X8 A0A182FF41 A0A023FA77 A0A2M4BW41 A0A182UHE1 A0A0L0BKR1 D6WFA2 Q5FVX2 A0A026W8F7 A0A336LXI2 A0A2J7QXC6 A0A1L8ECQ8 A0A2M3ZJG7 A0A1L8EBM6 Q6DJN9 F7A1E4 A0A2G8K236 A0A0P4VRY8 A0A1L8HYS5 A0A1L8ECT5 A0A1I8NQW8 A0A0V0G5D1 A0A0N8DC99 W8C3I0 A0A0N8BJS9 A0A0P6I897 A0A067QR58 E9FVF3 A0A1L8EA07 A0A182SN43 A0A0P5T9G6 A0A087UTH3 A0A1I8N4U5 A0A0P5JEB6 A0A1A9WBP1 A0A0N8CAM7 A0A0P5UCX3 A0A2P6K321 A0A2C9JMD9 A0A1A9VN14 A0A0P5ULN5 A0A0K8SGQ3 A0A0M3QVW0 A0A1A9ZMI0 D3TNI9 A0A3S1BUX9 B4IX02 A0A2S2NMB6 A0A0A9Z575 A0A0N8EGT2 A0A0P6BMU8 A0A0A1XNH2 B4KWZ6 A0A2B4S7B5 A0A0K8WF41 B4HUD8 T1KNI7 B4LFQ3
Pubmed
19121390
26354079
20074338
22118469
23691247
25244985
+ More
20966253 12364791 14747013 17210077 24438588 17510324 26483478 18362917 19820115 24945155 20920257 23761445 25474469 26108605 24508170 30249741 20431018 29023486 27129103 27762356 24495485 24845553 21292972 25315136 15562597 20353571 17994087 25401762 25830018
20966253 12364791 14747013 17210077 24438588 17510324 26483478 18362917 19820115 24945155 20920257 23761445 25474469 26108605 24508170 30249741 20431018 29023486 27129103 27762356 24495485 24845553 21292972 25315136 15562597 20353571 17994087 25401762 25830018
EMBL
BABH01035219
NWSH01001177
PCG72239.1
KQ459590
KPI97368.1
EZ407244
+ More
ACX53801.1 KQ461190 KPJ07176.1 ODYU01003588 SOQ42549.1 BABH01012008 BABH01012009 RSAL01000414 RVE41830.1 AGBW02013918 OWR42562.1 AK417828 BAN21043.1 APCN01000204 GANO01001366 JAB58505.1 AAAB01008807 EAA04619.4 GFDL01007282 JAV27763.1 AXCM01005343 ATLV01024580 KE525352 KFB51692.1 CH477380 EAT42217.1 LJIG01022786 KRT78724.1 JXUM01020768 KQ560581 KXJ81809.1 JXUM01043910 KQ561380 KXJ78774.1 GEDC01027926 JAS09372.1 DS232163 EDS36476.1 KQ971319 EFA00284.1 AXCN02000379 GECZ01022730 JAS47039.1 GAPW01003678 JAC09920.1 ADMH02000549 ETN65951.1 CVRI01000039 CRK94633.1 GGFM01005125 MBW25876.1 GGFJ01008134 MBW57275.1 GEBQ01021535 JAT18442.1 GBBI01000326 JAC18386.1 GGFJ01008138 MBW57279.1 JRES01001708 KNC20675.1 EFA00285.2 BC089728 AAH89728.1 KK107403 QOIP01000011 EZA51309.1 RLU16716.1 UFQS01000164 UFQT01000164 SSX00677.1 SSX21057.1 NEVH01009382 PNF33231.1 GFDG01002271 JAV16528.1 GGFM01007891 MBW28642.1 GFDG01002642 JAV16157.1 BC075136 AAH75136.1 AAMC01116011 MRZV01000964 PIK42067.1 GDKW01002757 JAI53838.1 CM004466 OCU01254.1 GFDG01002272 JAV16527.1 GECL01002949 JAP03175.1 GDIP01047855 LRGB01000024 JAM55860.1 KZS21785.1 GAMC01002737 JAC03819.1 GDIQ01170827 JAK80898.1 GDIQ01033977 JAN60760.1 KK853119 KDR11131.1 GL732525 EFX88544.1 GFDG01003238 JAV15561.1 GDIP01130750 JAL72964.1 KK121532 KFM80662.1 GDIQ01200808 JAK50917.1 GDIQ01098443 JAL53283.1 GDIP01132105 JAL71609.1 MWRG01034236 PRD20721.1 GDIP01111209 JAL92505.1 GBRD01013862 GBRD01013861 JAG51965.1 CP012525 ALC43079.1 CCAG010020395 EZ422991 ADD19267.1 RQTK01000001 RUS92170.1 CH916366 EDV96308.1 GGMR01005696 MBY18315.1 GBHO01003177 GBHO01003174 JAG40427.1 JAG40430.1 GDIQ01028212 JAN66525.1 GDIQ01241351 GDIP01027309 JAK10374.1 JAM76406.1 GBXI01001443 JAD12849.1 CH933809 EDW18617.1 LSMT01000163 PFX24979.1 GDHF01002645 JAI49669.1 CH480817 EDW50559.1 CAEY01000277 CH940647 EDW69280.1
ACX53801.1 KQ461190 KPJ07176.1 ODYU01003588 SOQ42549.1 BABH01012008 BABH01012009 RSAL01000414 RVE41830.1 AGBW02013918 OWR42562.1 AK417828 BAN21043.1 APCN01000204 GANO01001366 JAB58505.1 AAAB01008807 EAA04619.4 GFDL01007282 JAV27763.1 AXCM01005343 ATLV01024580 KE525352 KFB51692.1 CH477380 EAT42217.1 LJIG01022786 KRT78724.1 JXUM01020768 KQ560581 KXJ81809.1 JXUM01043910 KQ561380 KXJ78774.1 GEDC01027926 JAS09372.1 DS232163 EDS36476.1 KQ971319 EFA00284.1 AXCN02000379 GECZ01022730 JAS47039.1 GAPW01003678 JAC09920.1 ADMH02000549 ETN65951.1 CVRI01000039 CRK94633.1 GGFM01005125 MBW25876.1 GGFJ01008134 MBW57275.1 GEBQ01021535 JAT18442.1 GBBI01000326 JAC18386.1 GGFJ01008138 MBW57279.1 JRES01001708 KNC20675.1 EFA00285.2 BC089728 AAH89728.1 KK107403 QOIP01000011 EZA51309.1 RLU16716.1 UFQS01000164 UFQT01000164 SSX00677.1 SSX21057.1 NEVH01009382 PNF33231.1 GFDG01002271 JAV16528.1 GGFM01007891 MBW28642.1 GFDG01002642 JAV16157.1 BC075136 AAH75136.1 AAMC01116011 MRZV01000964 PIK42067.1 GDKW01002757 JAI53838.1 CM004466 OCU01254.1 GFDG01002272 JAV16527.1 GECL01002949 JAP03175.1 GDIP01047855 LRGB01000024 JAM55860.1 KZS21785.1 GAMC01002737 JAC03819.1 GDIQ01170827 JAK80898.1 GDIQ01033977 JAN60760.1 KK853119 KDR11131.1 GL732525 EFX88544.1 GFDG01003238 JAV15561.1 GDIP01130750 JAL72964.1 KK121532 KFM80662.1 GDIQ01200808 JAK50917.1 GDIQ01098443 JAL53283.1 GDIP01132105 JAL71609.1 MWRG01034236 PRD20721.1 GDIP01111209 JAL92505.1 GBRD01013862 GBRD01013861 JAG51965.1 CP012525 ALC43079.1 CCAG010020395 EZ422991 ADD19267.1 RQTK01000001 RUS92170.1 CH916366 EDV96308.1 GGMR01005696 MBY18315.1 GBHO01003177 GBHO01003174 JAG40427.1 JAG40430.1 GDIQ01028212 JAN66525.1 GDIQ01241351 GDIP01027309 JAK10374.1 JAM76406.1 GBXI01001443 JAD12849.1 CH933809 EDW18617.1 LSMT01000163 PFX24979.1 GDHF01002645 JAI49669.1 CH480817 EDW50559.1 CAEY01000277 CH940647 EDW69280.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000076407 UP000075840 UP000076408 UP000075903 UP000075920 UP000075882 UP000075900 UP000007062 UP000075883 UP000030765 UP000075884 UP000075885 UP000075880 UP000008820 UP000069940 UP000249989 UP000002320 UP000007266 UP000075886 UP000075881 UP000000673 UP000183832 UP000079169 UP000069272 UP000075902 UP000037069 UP000053097 UP000279307 UP000235965 UP000008143 UP000230750 UP000186698 UP000095300 UP000076858 UP000027135 UP000000305 UP000075901 UP000054359 UP000095301 UP000091820 UP000076420 UP000078200 UP000092553 UP000092445 UP000092444 UP000271974 UP000001070 UP000009192 UP000225706 UP000001292 UP000015104 UP000008792
UP000076407 UP000075840 UP000076408 UP000075903 UP000075920 UP000075882 UP000075900 UP000007062 UP000075883 UP000030765 UP000075884 UP000075885 UP000075880 UP000008820 UP000069940 UP000249989 UP000002320 UP000007266 UP000075886 UP000075881 UP000000673 UP000183832 UP000079169 UP000069272 UP000075902 UP000037069 UP000053097 UP000279307 UP000235965 UP000008143 UP000230750 UP000186698 UP000095300 UP000076858 UP000027135 UP000000305 UP000075901 UP000054359 UP000095301 UP000091820 UP000076420 UP000078200 UP000092553 UP000092445 UP000092444 UP000271974 UP000001070 UP000009192 UP000225706 UP000001292 UP000015104 UP000008792
Pfam
PF00106 adh_short
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JNK4
A0A2A4JK89
A0A194PWI4
D2SNW0
A0A194QUE6
A0A2H1VNY5
+ More
H9JQH9 A0A3S2L990 A0A212EM40 R4WDM8 A0A182X3R6 A0A182I3A3 A0A182XYV9 U5EWL2 A0A182VMY9 A0A182WJ87 A0A182KRB2 A0A182RSN1 Q7QJE4 A0A1Q3FJU4 A0A182MEK3 A0A084WN98 A0A1S4FCV9 A0A182NHV8 A0A182PQF5 A0A182IN68 Q176Z2 A0A0T6AUL9 A0A182GXS7 A0A182G5T3 A0A1B6C7C6 B0WXE6 D6WFA3 A0A182QY63 A0A182JV87 A0A1B6F9X4 A0A023EL45 W5JPB4 A0A1J1I894 A0A2M3ZBF2 A0A1S4ECP8 A0A2M4BW22 A0A1B6L3X8 A0A182FF41 A0A023FA77 A0A2M4BW41 A0A182UHE1 A0A0L0BKR1 D6WFA2 Q5FVX2 A0A026W8F7 A0A336LXI2 A0A2J7QXC6 A0A1L8ECQ8 A0A2M3ZJG7 A0A1L8EBM6 Q6DJN9 F7A1E4 A0A2G8K236 A0A0P4VRY8 A0A1L8HYS5 A0A1L8ECT5 A0A1I8NQW8 A0A0V0G5D1 A0A0N8DC99 W8C3I0 A0A0N8BJS9 A0A0P6I897 A0A067QR58 E9FVF3 A0A1L8EA07 A0A182SN43 A0A0P5T9G6 A0A087UTH3 A0A1I8N4U5 A0A0P5JEB6 A0A1A9WBP1 A0A0N8CAM7 A0A0P5UCX3 A0A2P6K321 A0A2C9JMD9 A0A1A9VN14 A0A0P5ULN5 A0A0K8SGQ3 A0A0M3QVW0 A0A1A9ZMI0 D3TNI9 A0A3S1BUX9 B4IX02 A0A2S2NMB6 A0A0A9Z575 A0A0N8EGT2 A0A0P6BMU8 A0A0A1XNH2 B4KWZ6 A0A2B4S7B5 A0A0K8WF41 B4HUD8 T1KNI7 B4LFQ3
H9JQH9 A0A3S2L990 A0A212EM40 R4WDM8 A0A182X3R6 A0A182I3A3 A0A182XYV9 U5EWL2 A0A182VMY9 A0A182WJ87 A0A182KRB2 A0A182RSN1 Q7QJE4 A0A1Q3FJU4 A0A182MEK3 A0A084WN98 A0A1S4FCV9 A0A182NHV8 A0A182PQF5 A0A182IN68 Q176Z2 A0A0T6AUL9 A0A182GXS7 A0A182G5T3 A0A1B6C7C6 B0WXE6 D6WFA3 A0A182QY63 A0A182JV87 A0A1B6F9X4 A0A023EL45 W5JPB4 A0A1J1I894 A0A2M3ZBF2 A0A1S4ECP8 A0A2M4BW22 A0A1B6L3X8 A0A182FF41 A0A023FA77 A0A2M4BW41 A0A182UHE1 A0A0L0BKR1 D6WFA2 Q5FVX2 A0A026W8F7 A0A336LXI2 A0A2J7QXC6 A0A1L8ECQ8 A0A2M3ZJG7 A0A1L8EBM6 Q6DJN9 F7A1E4 A0A2G8K236 A0A0P4VRY8 A0A1L8HYS5 A0A1L8ECT5 A0A1I8NQW8 A0A0V0G5D1 A0A0N8DC99 W8C3I0 A0A0N8BJS9 A0A0P6I897 A0A067QR58 E9FVF3 A0A1L8EA07 A0A182SN43 A0A0P5T9G6 A0A087UTH3 A0A1I8N4U5 A0A0P5JEB6 A0A1A9WBP1 A0A0N8CAM7 A0A0P5UCX3 A0A2P6K321 A0A2C9JMD9 A0A1A9VN14 A0A0P5ULN5 A0A0K8SGQ3 A0A0M3QVW0 A0A1A9ZMI0 D3TNI9 A0A3S1BUX9 B4IX02 A0A2S2NMB6 A0A0A9Z575 A0A0N8EGT2 A0A0P6BMU8 A0A0A1XNH2 B4KWZ6 A0A2B4S7B5 A0A0K8WF41 B4HUD8 T1KNI7 B4LFQ3
PDB
3O4R
E-value=7.14492e-63,
Score=608
Ontologies
PATHWAY
PANTHER
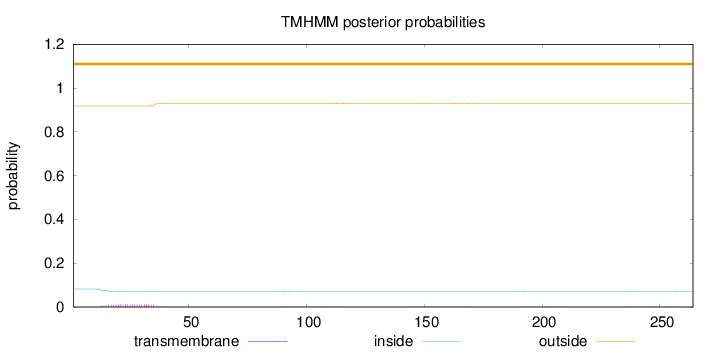
Topology
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28155
Exp number, first 60 AAs:
0.23156
Total prob of N-in:
0.08192
outside
1 - 264
Population Genetic Test Statistics
Pi
0.195287
Theta
1.763461
Tajima's D
-2.052478
CLR
946.93047
CSRT
0.00394980250987451
Interpretation
Possibly Positive selection
