Gene
KWMTBOMO14236
Pre Gene Modal
BGIBMGA011109
Annotation
PREDICTED:_23_kDa_integral_membrane_protein-like_isoform_X3_[Bombyx_mori]
Full name
Tetraspanin
Location in the cell
PlasmaMembrane Reliability : 4.984
Sequence
CDS
ATGTGTTGCCCCGAGTTTATAGCTAAATATGTACTCTTCATCGCCAACCTTGTTTTTTCGTTGGCAGGTCTAGCTATTATACGTCTTGGAGTGGCCGTCTTAAGAAATCTACGGGATCTTCAAGATATTCTACCAGTCAACGCTTTGCCAATAGGGATAATCGTCCTCGGATGCATCATATTCATCATTGCATTCCTAGCGTGCTGTGGAGCGATAAAGGAATCAAGATGTATGCTGATAACCTACTCCATATTCATGGTGATACTGGTGGCCGTGAAGATCTACCTGGCTATCGTGGTATTCGGTTTTCTGTCCGACGTCACCAGCACAATAACCAGCTGGGTGACCACAGCGTTCAACACGAGCAGCTTAAGAGACGTATATCACGTCATGGAAGCTCTATTCAATTGCTGTGGAACAACCGGCCCCTCTTCGTACGACGGTATCTTGTCACAGCTGCCGCCCTCGTGCTGCGCCAGCCCCGTTGACAACACCTTCTATGCCCCCAACGCGTTCCCCGGCTGCACCACTCGACTGATTGATTACTTCGATACCTTCGGTAGGGCCATCGGCAGCGTTCTGATCATTATCATATTTCTTGAGTTCATCTCGATCATCTTCGCTTGGTTCCTGTCGCATTCTTATTCGCAGAAACGGCGCGGGAATATAGCATAA
Protein
MCCPEFIAKYVLFIANLVFSLAGLAIIRLGVAVLRNLRDLQDILPVNALPIGIIVLGCIIFIIAFLACCGAIKESRCMLITYSIFMVILVAVKIYLAIVVFGFLSDVTSTITSWVTTAFNTSSLRDVYHVMEALFNCCGTTGPSSYDGILSQLPPSCCASPVDNTFYAPNAFPGCTTRLIDYFDTFGRAIGSVLIIIIFLEFISIIFAWFLSHSYSQKRRGNIA
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Uniprot
H9JNK5
A0A2A4K689
A0A2W1BPW8
A0A3S2NM92
A0A1E1W0A4
S4PF79
+ More
A0A2A4K899 A0A3S2LT60 Q1HQ39 A0A212EGN7 A0A194RHI5 I4DJ82 A0A194QGK8 A0A1L8DKH3 A0A2A4JXL7 I4DPV3 A0A1L8DJJ6 Q9NB08 V5I9N6 I4DMY0 A0A0G4AJH1 A0A1E1WV10 A0A1E1W4P8 A0A1B0GKM0 S4PWL6 E0VMX8 A0A0C9Q5P6 A0A084WER4 A0A182I2Q6 A7URL1 Q7PTA7 A0A194Q3K1 A0A212EWY8 R4V3T2 A0A182N923 N6U6Y0 A0A182Q6M3 A0A067QYE5 A0A182YML0 A0A0A1X8C6 A0A1I8PEJ6 A0A2M4CTQ4 A0A2M4CHV5 A0A2M4CV63 W8BXN6 A0A2M4BYW8 A0A2M4A0S0 A0A2M4A0K4 A0A2M4CQ62 A0A224XJR8 T1GNT4 A0A2J7QP49 V5G3W6 W8CAJ3 B4M9T6 A0A1I8NGG4 A0A1S4G0M8 A0A1S4G0S2 A0A139WBP1 Q7K1I4 A0A1I8QAH3 A0A182IP56 A0A1L8EG93 B3MHM8 A0A1W4V1F2 A0A1J1IZR8 A0A034V4M1 A0A023EK90 A0A1W4WL05 A0A023EK84 B4P1B8 T1E2B6 Q16HX6 Q16HX5 B4MPV5 B4KF00 Q292W0 A0A182TSU7 B4P1C0 A0A194QWI4 A0A1J1IM26 A0A194Q3I5 A0A0T6AV57 A0A182KQN6 A0A034WBD7 A0A0C9R4C5 A0A1L8EFL5 A0A0M4E1J6 Q292W2 A0A1I8MN26
A0A2A4K899 A0A3S2LT60 Q1HQ39 A0A212EGN7 A0A194RHI5 I4DJ82 A0A194QGK8 A0A1L8DKH3 A0A2A4JXL7 I4DPV3 A0A1L8DJJ6 Q9NB08 V5I9N6 I4DMY0 A0A0G4AJH1 A0A1E1WV10 A0A1E1W4P8 A0A1B0GKM0 S4PWL6 E0VMX8 A0A0C9Q5P6 A0A084WER4 A0A182I2Q6 A7URL1 Q7PTA7 A0A194Q3K1 A0A212EWY8 R4V3T2 A0A182N923 N6U6Y0 A0A182Q6M3 A0A067QYE5 A0A182YML0 A0A0A1X8C6 A0A1I8PEJ6 A0A2M4CTQ4 A0A2M4CHV5 A0A2M4CV63 W8BXN6 A0A2M4BYW8 A0A2M4A0S0 A0A2M4A0K4 A0A2M4CQ62 A0A224XJR8 T1GNT4 A0A2J7QP49 V5G3W6 W8CAJ3 B4M9T6 A0A1I8NGG4 A0A1S4G0M8 A0A1S4G0S2 A0A139WBP1 Q7K1I4 A0A1I8QAH3 A0A182IP56 A0A1L8EG93 B3MHM8 A0A1W4V1F2 A0A1J1IZR8 A0A034V4M1 A0A023EK90 A0A1W4WL05 A0A023EK84 B4P1B8 T1E2B6 Q16HX6 Q16HX5 B4MPV5 B4KF00 Q292W0 A0A182TSU7 B4P1C0 A0A194QWI4 A0A1J1IM26 A0A194Q3I5 A0A0T6AV57 A0A182KQN6 A0A034WBD7 A0A0C9R4C5 A0A1L8EFL5 A0A0M4E1J6 Q292W2 A0A1I8MN26
Pubmed
19121390
28756777
23622113
22118469
26354079
22651552
+ More
11122467 20566863 24438588 12364791 14747013 17210077 23537049 24845553 25244985 25830018 24495485 17994087 25315136 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 24945155 17550304 24330624 17510324 15632085 20966253
11122467 20566863 24438588 12364791 14747013 17210077 23537049 24845553 25244985 25830018 24495485 17994087 25315136 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 24945155 17550304 24330624 17510324 15632085 20966253
EMBL
BABH01035216
BABH01035217
BABH01035218
NWSH01000112
PCG79438.1
KZ149990
+ More
PZC75605.1 RSAL01000264 RVE43295.1 GDQN01010636 JAT80418.1 GAIX01004167 JAA88393.1 NWSH01000032 PCG80495.1 RVE43294.1 DQ443213 ABF51302.1 AGBW02015051 OWR40653.1 KQ460154 KPJ17288.1 AK401350 BAM17972.1 KQ458880 KPJ04637.1 GFDF01007244 JAV06840.1 NWSH01000388 PCG76761.1 AK403798 BAM19943.1 GFDF01007446 JAV06638.1 AF274023 AAF90149.1 GALX01002620 JAB65846.1 AK402648 BAM19270.1 KP999984 AKM70275.1 GDQN01000423 JAT90631.1 GDQN01010381 GDQN01009120 JAT80673.1 JAT81934.1 AJWK01030694 AJWK01030695 GAIX01006573 JAA85987.1 DS235327 EEB14734.1 GBYB01009418 JAG79185.1 ATLV01023246 KE525341 KFB48708.1 APCN01000178 AAAB01008807 EDO64653.1 EAA04202.4 KQ459472 KPJ00118.1 AGBW02011867 OWR45999.1 KC571964 AGM32463.1 APGK01047299 APGK01047300 KB741077 KB631602 ENN74342.1 ERL84575.1 AXCN02000540 KK853109 KDR11258.1 GBXI01007136 JAD07156.1 GGFL01004539 MBW68717.1 GGFL01000705 MBW64883.1 GGFL01004540 MBW68718.1 GAMC01002483 JAC04073.1 GGFJ01009053 MBW58194.1 GGFK01001038 MBW34359.1 GGFK01001025 MBW34346.1 GGFL01003304 MBW67482.1 GFTR01003691 JAW12735.1 CAQQ02119555 CAQQ02119556 CAQQ02119557 CAQQ02119558 CAQQ02119559 CAQQ02119560 NEVH01012097 PNF30360.1 GALX01003741 JAB64725.1 GAMC01005696 GAMC01005695 JAC00860.1 CH940654 EDW57962.1 KQ971372 KYB25323.1 AE013599 AF274014 AY069049 AAF57420.2 AAF90140.1 AAL39194.1 GFDG01001052 JAV17747.1 CH902619 EDV35864.1 CVRI01000065 CRL05701.1 GAKP01020761 JAC38191.1 GAPW01004058 JAC09540.1 GAPW01004057 JAC09541.1 CM000157 EDW89120.1 GALA01000879 JAA93973.1 CH478132 EAT33857.1 EAT33856.1 CH963849 EDW74144.1 CH933807 EDW11969.1 CM000071 EAL24751.1 EDW89122.1 KQ461175 KPJ07931.1 CVRI01000054 CRL00126.1 KPJ00103.1 LJIG01022738 KRT78969.1 GAKP01007859 JAC51093.1 GBYB01002740 JAG72507.1 GFDG01001394 JAV17405.1 CP012523 ALC39254.1 EAL24750.1
PZC75605.1 RSAL01000264 RVE43295.1 GDQN01010636 JAT80418.1 GAIX01004167 JAA88393.1 NWSH01000032 PCG80495.1 RVE43294.1 DQ443213 ABF51302.1 AGBW02015051 OWR40653.1 KQ460154 KPJ17288.1 AK401350 BAM17972.1 KQ458880 KPJ04637.1 GFDF01007244 JAV06840.1 NWSH01000388 PCG76761.1 AK403798 BAM19943.1 GFDF01007446 JAV06638.1 AF274023 AAF90149.1 GALX01002620 JAB65846.1 AK402648 BAM19270.1 KP999984 AKM70275.1 GDQN01000423 JAT90631.1 GDQN01010381 GDQN01009120 JAT80673.1 JAT81934.1 AJWK01030694 AJWK01030695 GAIX01006573 JAA85987.1 DS235327 EEB14734.1 GBYB01009418 JAG79185.1 ATLV01023246 KE525341 KFB48708.1 APCN01000178 AAAB01008807 EDO64653.1 EAA04202.4 KQ459472 KPJ00118.1 AGBW02011867 OWR45999.1 KC571964 AGM32463.1 APGK01047299 APGK01047300 KB741077 KB631602 ENN74342.1 ERL84575.1 AXCN02000540 KK853109 KDR11258.1 GBXI01007136 JAD07156.1 GGFL01004539 MBW68717.1 GGFL01000705 MBW64883.1 GGFL01004540 MBW68718.1 GAMC01002483 JAC04073.1 GGFJ01009053 MBW58194.1 GGFK01001038 MBW34359.1 GGFK01001025 MBW34346.1 GGFL01003304 MBW67482.1 GFTR01003691 JAW12735.1 CAQQ02119555 CAQQ02119556 CAQQ02119557 CAQQ02119558 CAQQ02119559 CAQQ02119560 NEVH01012097 PNF30360.1 GALX01003741 JAB64725.1 GAMC01005696 GAMC01005695 JAC00860.1 CH940654 EDW57962.1 KQ971372 KYB25323.1 AE013599 AF274014 AY069049 AAF57420.2 AAF90140.1 AAL39194.1 GFDG01001052 JAV17747.1 CH902619 EDV35864.1 CVRI01000065 CRL05701.1 GAKP01020761 JAC38191.1 GAPW01004058 JAC09540.1 GAPW01004057 JAC09541.1 CM000157 EDW89120.1 GALA01000879 JAA93973.1 CH478132 EAT33857.1 EAT33856.1 CH963849 EDW74144.1 CH933807 EDW11969.1 CM000071 EAL24751.1 EDW89122.1 KQ461175 KPJ07931.1 CVRI01000054 CRL00126.1 KPJ00103.1 LJIG01022738 KRT78969.1 GAKP01007859 JAC51093.1 GBYB01002740 JAG72507.1 GFDG01001394 JAV17405.1 CP012523 ALC39254.1 EAL24750.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000092461 UP000009046 UP000030765 UP000075840 UP000007062 UP000075884 UP000019118 UP000030742 UP000075886 UP000027135 UP000076408 UP000095300 UP000015102 UP000235965 UP000008792 UP000095301 UP000007266 UP000000803 UP000075880 UP000007801 UP000192221 UP000183832 UP000192223 UP000002282 UP000008820 UP000007798 UP000009192 UP000001819 UP000075902 UP000075882 UP000092553
UP000092461 UP000009046 UP000030765 UP000075840 UP000007062 UP000075884 UP000019118 UP000030742 UP000075886 UP000027135 UP000076408 UP000095300 UP000015102 UP000235965 UP000008792 UP000095301 UP000007266 UP000000803 UP000075880 UP000007801 UP000192221 UP000183832 UP000192223 UP000002282 UP000008820 UP000007798 UP000009192 UP000001819 UP000075902 UP000075882 UP000092553
Pfam
PF00335 Tetraspanin
Interpro
Gene 3D
ProteinModelPortal
H9JNK5
A0A2A4K689
A0A2W1BPW8
A0A3S2NM92
A0A1E1W0A4
S4PF79
+ More
A0A2A4K899 A0A3S2LT60 Q1HQ39 A0A212EGN7 A0A194RHI5 I4DJ82 A0A194QGK8 A0A1L8DKH3 A0A2A4JXL7 I4DPV3 A0A1L8DJJ6 Q9NB08 V5I9N6 I4DMY0 A0A0G4AJH1 A0A1E1WV10 A0A1E1W4P8 A0A1B0GKM0 S4PWL6 E0VMX8 A0A0C9Q5P6 A0A084WER4 A0A182I2Q6 A7URL1 Q7PTA7 A0A194Q3K1 A0A212EWY8 R4V3T2 A0A182N923 N6U6Y0 A0A182Q6M3 A0A067QYE5 A0A182YML0 A0A0A1X8C6 A0A1I8PEJ6 A0A2M4CTQ4 A0A2M4CHV5 A0A2M4CV63 W8BXN6 A0A2M4BYW8 A0A2M4A0S0 A0A2M4A0K4 A0A2M4CQ62 A0A224XJR8 T1GNT4 A0A2J7QP49 V5G3W6 W8CAJ3 B4M9T6 A0A1I8NGG4 A0A1S4G0M8 A0A1S4G0S2 A0A139WBP1 Q7K1I4 A0A1I8QAH3 A0A182IP56 A0A1L8EG93 B3MHM8 A0A1W4V1F2 A0A1J1IZR8 A0A034V4M1 A0A023EK90 A0A1W4WL05 A0A023EK84 B4P1B8 T1E2B6 Q16HX6 Q16HX5 B4MPV5 B4KF00 Q292W0 A0A182TSU7 B4P1C0 A0A194QWI4 A0A1J1IM26 A0A194Q3I5 A0A0T6AV57 A0A182KQN6 A0A034WBD7 A0A0C9R4C5 A0A1L8EFL5 A0A0M4E1J6 Q292W2 A0A1I8MN26
A0A2A4K899 A0A3S2LT60 Q1HQ39 A0A212EGN7 A0A194RHI5 I4DJ82 A0A194QGK8 A0A1L8DKH3 A0A2A4JXL7 I4DPV3 A0A1L8DJJ6 Q9NB08 V5I9N6 I4DMY0 A0A0G4AJH1 A0A1E1WV10 A0A1E1W4P8 A0A1B0GKM0 S4PWL6 E0VMX8 A0A0C9Q5P6 A0A084WER4 A0A182I2Q6 A7URL1 Q7PTA7 A0A194Q3K1 A0A212EWY8 R4V3T2 A0A182N923 N6U6Y0 A0A182Q6M3 A0A067QYE5 A0A182YML0 A0A0A1X8C6 A0A1I8PEJ6 A0A2M4CTQ4 A0A2M4CHV5 A0A2M4CV63 W8BXN6 A0A2M4BYW8 A0A2M4A0S0 A0A2M4A0K4 A0A2M4CQ62 A0A224XJR8 T1GNT4 A0A2J7QP49 V5G3W6 W8CAJ3 B4M9T6 A0A1I8NGG4 A0A1S4G0M8 A0A1S4G0S2 A0A139WBP1 Q7K1I4 A0A1I8QAH3 A0A182IP56 A0A1L8EG93 B3MHM8 A0A1W4V1F2 A0A1J1IZR8 A0A034V4M1 A0A023EK90 A0A1W4WL05 A0A023EK84 B4P1B8 T1E2B6 Q16HX6 Q16HX5 B4MPV5 B4KF00 Q292W0 A0A182TSU7 B4P1C0 A0A194QWI4 A0A1J1IM26 A0A194Q3I5 A0A0T6AV57 A0A182KQN6 A0A034WBD7 A0A0C9R4C5 A0A1L8EFL5 A0A0M4E1J6 Q292W2 A0A1I8MN26
Ontologies
GO
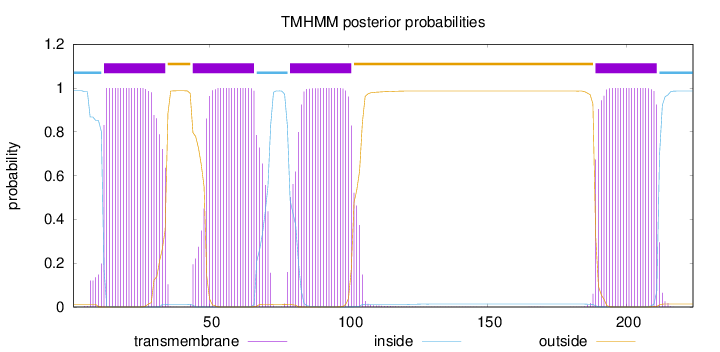
Topology
Subcellular location
Membrane
Length:
224
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
90.95622
Exp number, first 60 AAs:
35.81924
Total prob of N-in:
0.98760
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 43
TMhelix
44 - 66
inside
67 - 78
TMhelix
79 - 101
outside
102 - 188
TMhelix
189 - 211
inside
212 - 224
Population Genetic Test Statistics
Pi
1.463428
Theta
4.855066
Tajima's D
-1.739907
CLR
298.821529
CSRT
0.0289985500724964
Interpretation
Possibly Positive selection
