Gene
KWMTBOMO14231
Pre Gene Modal
BGIBMGA011153
Annotation
PREDICTED:_scavenger_receptor_class_B_member_1_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.932
Sequence
CDS
ATGTCTAGTGGCTTGGCGCTGATCCTGGCATTGGTGAAGTCCATAGGCGACGGTAATCATCAGGCGAACCGGTTCGTGCTCGCATCAGCAGTCATCCTACTCCTCCTAGGCATAGTCATTGCCATATTCCTCGGGACTTGGATTCGACTCTTCATAGACCATGAGCTGGTGCTTCGCCCAGGTTCGATGACGTTCGAGTGGTGGGCGCGGCCTCCGGTGCGGCCCTTCGTCAAGGTCTACGTGTACAACGTGACCAACGCCGATGAGTTCCTCAACAACGGCTCCAAGCCGATACTGGACGAGCTCGGACCATATGTTTACTCTGAGGAGTGGGAGAAGATCAACATAACAGACAACGAGAATGGCACGCTGTCCTTCCACTACAAGAGGACGTACACCTTCCAGCCGGAGATGAGCAACGGGATGGACGACGATGCGGTCGTCGTGCCCAACATACCGATGCTGAGCGCGACATCTCAATCGAAGCACGCCGCTAGGTTCCTGCGTCTCGCCATGGCTTCCATCATGGACATATTGAAGATCAAGCCTTTTGTCGAGGTGTCCGTCGGTCAGCTGCTCTGGGGCTATGAAGATCCGCTCCTGAAGTTGGCCAAGGACGTCGTGCCCAAGGAGCAGAAGTTGCCTTACGAGGAATTCGGACTTTTTTATGGCAAGAACGGCACCTCTCCGGACGTGGTGACGATGTTCACTGGTGCTACGGACATGACCCGCTACGGCATCGTGGACCGCTACAACATGAGGGACCGGCTGCCCCACTGGACCACGGACCAGTGCAACAGCGTCGCCGGCTCCGACGGGTCCATCTTCCCGCCGCACATCACCGCGAACGACACGCTCCGGGTCTACGACAAGGACCTCTGCCGATTGCTTCCGCTGAGGTATCTCGAAACGGTGGAATCTTCAGCCGGAGTCGAAGGCTATAGGTTCACTCCCCCTGAGGATGTCTTCGCTAACGACGAGCACAATAAGTGCTTCTGTCCTGCCGGTCCCCCCTGCGCCCCCGACGGACTCTTCAACGTCTCCCTCTGCCAATACGATTCTCCCATAATGCTATCGTTCCCTCACTTCTACTTGGCGGACCCGAGCCTGCGGGAGGCAGTCGAGGGCATCTCACCACCAGTACCAGAGAAGCACAGACTGTACATCGACGTGCAGCCCGAGATGGGTATAGCAATGCGCGCTCGAGCTAGGATCCAGATTAACCTGGCCGTCTCCCAGGTCTTGGACATCAAGCAGGTGGCCAACTTCCCAGATATCGTCTTCCCCATATTGTGGTTCGAAGAGGGTATAGACGAGCTGCCGGAGCGGGTGTCGTCGCTGCTGCGGCTGGCGACGCGCGTGCCGGGCGTGGCGCGGGCGGCGCTGGCGGGCGGGCTGTGCGCGCTGGGCGCCGCGCTCGTGCTGCTCGCCGTCACCTGCCTCATCAGATCGTCTCACAGACAAAGCACTCTCAGGCTGGAGGCTCACGTTGTCGCGAAGCCGCCTCCACCCAAAACACCGAGCAAAGAAAGCGCCTACGAACTGAACAGCCGGAGATAA
Protein
MSSGLALILALVKSIGDGNHQANRFVLASAVILLLLGIVIAIFLGTWIRLFIDHELVLRPGSMTFEWWARPPVRPFVKVYVYNVTNADEFLNNGSKPILDELGPYVYSEEWEKINITDNENGTLSFHYKRTYTFQPEMSNGMDDDAVVVPNIPMLSATSQSKHAARFLRLAMASIMDILKIKPFVEVSVGQLLWGYEDPLLKLAKDVVPKEQKLPYEEFGLFYGKNGTSPDVVTMFTGATDMTRYGIVDRYNMRDRLPHWTTDQCNSVAGSDGSIFPPHITANDTLRVYDKDLCRLLPLRYLETVESSAGVEGYRFTPPEDVFANDEHNKCFCPAGPPCAPDGLFNVSLCQYDSPIMLSFPHFYLADPSLREAVEGISPPVPEKHRLYIDVQPEMGIAMRARARIQINLAVSQVLDIKQVANFPDIVFPILWFEEGIDELPERVSSLLRLATRVPGVARAALAGGLCALGAALVLLAVTCLIRSSHRQSTLRLEAHVVAKPPPPKTPSKESAYELNSRR
Summary
Similarity
Belongs to the CD36 family.
Uniprot
H9JNP9
A0A194PSG6
A0A2A4JXP2
A0A2A4JWL0
A0A2W1BS53
A0A212ELF4
+ More
S4PIN9 A0A0A9Z111 A0A146L3X1 A0A0A9YZJ9 A0A1L8DJY2 A0A1L8DJU8 A0A1L8DJU1 A0A1Q3FZ01 A0A2M4CTR7 A0A1B6CLY9 A0A182QT15 A0A2M4BKE1 A0A182YBW3 A0A182WMJ8 A0A182FCS4 A0A023ETI9 A0A2M4AF82 E0W153 A0A182J8X5 A0A182P0K9 A0A182NV30 A0A182K3R0 A0A182LHW8 A0A2C9H6J5 A0A182RQG1 A0A084VUD8 A7UTD1 Q17PS2 A0A182WS42 A0A182U010 A0A182VL64 Q7PHM8 A0A182HXT8 D2K2F5 A0A0L0CBL7 A0A1B0A1R7 A0A224XN62 A0A1B0BQW7 A0A1A9YD57 U5ET11 A0A067R9W5 A0A336MNJ0 A0A1I8PMR0 A0A0A1WD10 T1PCA8 A0A034V7C7 A0A0K8V6P0 T1IBV5 A0A0J9RKW7 B4QD55 A0A0J9RL49 A0A0B4LGP3 Q9W0X0 Q7KVF1 W8AZM4 Q8IGY0 A0A0R3NRJ3 A0A0Q5VNZ8 B4GDA2 B4PBY8 Q292L3 B3NR92 A0A0Q5VYK1 A0A1W4V0J0 A0A0Q9WFT2 A0A3B0J111 B4MNU8 Q8SZV3 A0A3B0JMG0 B4LL12 A0A0L7LTQ3 D2A3S6 A0A0P8XQ07 B3MCD0 B4J9L8 A0A0M4EWV8 A0A1B6L2A4 A0A1B6ES61 A0A0Q9XHK8 B4KQT1 A0A1A9WI71 E9IFP2 A0A1Y1M5E8 A0A195FWH9 A0A194RIS4 A0A3Q9ELP8 A0A026WY03 A0A3L8DGW4 A0A0C9R2F4 Q24336 A0A2H1V4H1 A0A1B0GC45 A0A0J7L9G5 A0A195DD19
S4PIN9 A0A0A9Z111 A0A146L3X1 A0A0A9YZJ9 A0A1L8DJY2 A0A1L8DJU8 A0A1L8DJU1 A0A1Q3FZ01 A0A2M4CTR7 A0A1B6CLY9 A0A182QT15 A0A2M4BKE1 A0A182YBW3 A0A182WMJ8 A0A182FCS4 A0A023ETI9 A0A2M4AF82 E0W153 A0A182J8X5 A0A182P0K9 A0A182NV30 A0A182K3R0 A0A182LHW8 A0A2C9H6J5 A0A182RQG1 A0A084VUD8 A7UTD1 Q17PS2 A0A182WS42 A0A182U010 A0A182VL64 Q7PHM8 A0A182HXT8 D2K2F5 A0A0L0CBL7 A0A1B0A1R7 A0A224XN62 A0A1B0BQW7 A0A1A9YD57 U5ET11 A0A067R9W5 A0A336MNJ0 A0A1I8PMR0 A0A0A1WD10 T1PCA8 A0A034V7C7 A0A0K8V6P0 T1IBV5 A0A0J9RKW7 B4QD55 A0A0J9RL49 A0A0B4LGP3 Q9W0X0 Q7KVF1 W8AZM4 Q8IGY0 A0A0R3NRJ3 A0A0Q5VNZ8 B4GDA2 B4PBY8 Q292L3 B3NR92 A0A0Q5VYK1 A0A1W4V0J0 A0A0Q9WFT2 A0A3B0J111 B4MNU8 Q8SZV3 A0A3B0JMG0 B4LL12 A0A0L7LTQ3 D2A3S6 A0A0P8XQ07 B3MCD0 B4J9L8 A0A0M4EWV8 A0A1B6L2A4 A0A1B6ES61 A0A0Q9XHK8 B4KQT1 A0A1A9WI71 E9IFP2 A0A1Y1M5E8 A0A195FWH9 A0A194RIS4 A0A3Q9ELP8 A0A026WY03 A0A3L8DGW4 A0A0C9R2F4 Q24336 A0A2H1V4H1 A0A1B0GC45 A0A0J7L9G5 A0A195DD19
Pubmed
19121390
26354079
28756777
22118469
23622113
25401762
+ More
26823975 25244985 24945155 20566863 20966253 24438588 12364791 14747013 17210077 17510324 20230829 26108605 24845553 25830018 25315136 25348373 22936249 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085 17550304 23185243 18057021 26227816 18362917 19820115 21282665 28004739 30542294 24508170 30249741 7693949
26823975 25244985 24945155 20566863 20966253 24438588 12364791 14747013 17210077 17510324 20230829 26108605 24845553 25830018 25315136 25348373 22936249 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085 17550304 23185243 18057021 26227816 18362917 19820115 21282665 28004739 30542294 24508170 30249741 7693949
EMBL
BABH01035208
KQ459593
KPI96381.1
NWSH01000441
PCG76404.1
PCG76405.1
+ More
KZ149990 PZC75600.1 AGBW02014062 OWR42330.1 GAIX01005330 JAA87230.1 GBHO01006566 GBHO01006565 GBRD01009533 JAG37038.1 JAG37039.1 JAG56291.1 GDHC01015425 JAQ03204.1 GBHO01006563 JAG37041.1 GFDF01007353 JAV06731.1 GFDF01007352 JAV06732.1 GFDF01007335 JAV06749.1 GFDL01002282 JAV32763.1 GGFL01004393 MBW68571.1 GEDC01022903 JAS14395.1 AXCN02001921 GGFJ01004302 MBW53443.1 GAPW01000970 GEHC01000812 JAC12628.1 JAV46833.1 GGFK01006128 MBW39449.1 DS235866 EEB19359.1 ATLV01016723 ATLV01016724 KE525103 KFB41582.1 AAAB01008905 EDO64039.1 CH477189 EAT48769.1 EAA09703.2 EAA44477.4 APCN01005348 GU175173 ACZ58351.1 JRES01000644 KNC29651.1 GFTR01006997 JAW09429.1 JXJN01018835 GANO01002985 JAB56886.1 KK852602 KDR20477.1 UFQT01001469 SSX30709.1 GBXI01017525 JAC96766.1 KA645573 KA649301 AFP60202.1 GAKP01020920 JAC38032.1 GDHF01032913 GDHF01018089 JAI19401.1 JAI34225.1 ACPB03004980 CM002911 KMY96501.1 CM000362 EDX08651.1 KMY96500.1 KMY96502.1 KMY96503.1 AE013599 AHN56668.1 AAM70803.2 AAF47309.1 AAM70804.1 AHN56667.1 GAMC01012270 JAB94285.1 BT001535 AAN71290.1 CM000071 KRT01621.1 CH954179 KQS63165.1 CH479181 EDW31578.1 CM000158 EDW92642.2 EAL24849.1 KRT01622.1 EDV57111.1 KQS63163.1 KQS63164.1 CH940648 KRF80260.1 OUUW01000001 SPP74475.1 CH963848 EDW73787.2 AY070494 AAL47965.1 SPP74476.1 EDW61819.1 KRF80261.1 JTDY01000119 KOB78739.1 KQ971348 EFA05670.1 CH902619 KPU76658.1 EDV37254.1 KPU76657.1 CH916367 EDW02525.1 CP012524 ALC42370.1 GEBQ01022202 GEBQ01012636 GEBQ01007898 JAT17775.1 JAT27341.1 JAT32079.1 GECZ01028983 JAS40786.1 CH933808 KRG04562.1 EDW09280.1 KRG04561.1 GL762880 EFZ20621.1 GEZM01041504 JAV80298.1 KQ981208 KYN44672.1 KQ460152 KPJ17344.1 MK049033 AZQ24974.1 KK107064 EZA60922.1 QOIP01000008 RLU19685.1 GBYB01001046 JAG70813.1 X73332 CAA51759.1 ODYU01000628 SOQ35733.1 CCAG010018987 LBMM01000192 KMR04532.1 KQ980989 KYN10727.1
KZ149990 PZC75600.1 AGBW02014062 OWR42330.1 GAIX01005330 JAA87230.1 GBHO01006566 GBHO01006565 GBRD01009533 JAG37038.1 JAG37039.1 JAG56291.1 GDHC01015425 JAQ03204.1 GBHO01006563 JAG37041.1 GFDF01007353 JAV06731.1 GFDF01007352 JAV06732.1 GFDF01007335 JAV06749.1 GFDL01002282 JAV32763.1 GGFL01004393 MBW68571.1 GEDC01022903 JAS14395.1 AXCN02001921 GGFJ01004302 MBW53443.1 GAPW01000970 GEHC01000812 JAC12628.1 JAV46833.1 GGFK01006128 MBW39449.1 DS235866 EEB19359.1 ATLV01016723 ATLV01016724 KE525103 KFB41582.1 AAAB01008905 EDO64039.1 CH477189 EAT48769.1 EAA09703.2 EAA44477.4 APCN01005348 GU175173 ACZ58351.1 JRES01000644 KNC29651.1 GFTR01006997 JAW09429.1 JXJN01018835 GANO01002985 JAB56886.1 KK852602 KDR20477.1 UFQT01001469 SSX30709.1 GBXI01017525 JAC96766.1 KA645573 KA649301 AFP60202.1 GAKP01020920 JAC38032.1 GDHF01032913 GDHF01018089 JAI19401.1 JAI34225.1 ACPB03004980 CM002911 KMY96501.1 CM000362 EDX08651.1 KMY96500.1 KMY96502.1 KMY96503.1 AE013599 AHN56668.1 AAM70803.2 AAF47309.1 AAM70804.1 AHN56667.1 GAMC01012270 JAB94285.1 BT001535 AAN71290.1 CM000071 KRT01621.1 CH954179 KQS63165.1 CH479181 EDW31578.1 CM000158 EDW92642.2 EAL24849.1 KRT01622.1 EDV57111.1 KQS63163.1 KQS63164.1 CH940648 KRF80260.1 OUUW01000001 SPP74475.1 CH963848 EDW73787.2 AY070494 AAL47965.1 SPP74476.1 EDW61819.1 KRF80261.1 JTDY01000119 KOB78739.1 KQ971348 EFA05670.1 CH902619 KPU76658.1 EDV37254.1 KPU76657.1 CH916367 EDW02525.1 CP012524 ALC42370.1 GEBQ01022202 GEBQ01012636 GEBQ01007898 JAT17775.1 JAT27341.1 JAT32079.1 GECZ01028983 JAS40786.1 CH933808 KRG04562.1 EDW09280.1 KRG04561.1 GL762880 EFZ20621.1 GEZM01041504 JAV80298.1 KQ981208 KYN44672.1 KQ460152 KPJ17344.1 MK049033 AZQ24974.1 KK107064 EZA60922.1 QOIP01000008 RLU19685.1 GBYB01001046 JAG70813.1 X73332 CAA51759.1 ODYU01000628 SOQ35733.1 CCAG010018987 LBMM01000192 KMR04532.1 KQ980989 KYN10727.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000075886
UP000076408
+ More
UP000075920 UP000069272 UP000009046 UP000075880 UP000075885 UP000075884 UP000075881 UP000075882 UP000076407 UP000075900 UP000030765 UP000007062 UP000008820 UP000075902 UP000075903 UP000075840 UP000037069 UP000092445 UP000092460 UP000092443 UP000027135 UP000095300 UP000095301 UP000015103 UP000000304 UP000000803 UP000001819 UP000008711 UP000008744 UP000002282 UP000192221 UP000008792 UP000268350 UP000007798 UP000037510 UP000007266 UP000007801 UP000001070 UP000092553 UP000009192 UP000091820 UP000078541 UP000053240 UP000053097 UP000279307 UP000092444 UP000036403 UP000078492
UP000075920 UP000069272 UP000009046 UP000075880 UP000075885 UP000075884 UP000075881 UP000075882 UP000076407 UP000075900 UP000030765 UP000007062 UP000008820 UP000075902 UP000075903 UP000075840 UP000037069 UP000092445 UP000092460 UP000092443 UP000027135 UP000095300 UP000095301 UP000015103 UP000000304 UP000000803 UP000001819 UP000008711 UP000008744 UP000002282 UP000192221 UP000008792 UP000268350 UP000007798 UP000037510 UP000007266 UP000007801 UP000001070 UP000092553 UP000009192 UP000091820 UP000078541 UP000053240 UP000053097 UP000279307 UP000092444 UP000036403 UP000078492
Pfam
PF01130 CD36
ProteinModelPortal
H9JNP9
A0A194PSG6
A0A2A4JXP2
A0A2A4JWL0
A0A2W1BS53
A0A212ELF4
+ More
S4PIN9 A0A0A9Z111 A0A146L3X1 A0A0A9YZJ9 A0A1L8DJY2 A0A1L8DJU8 A0A1L8DJU1 A0A1Q3FZ01 A0A2M4CTR7 A0A1B6CLY9 A0A182QT15 A0A2M4BKE1 A0A182YBW3 A0A182WMJ8 A0A182FCS4 A0A023ETI9 A0A2M4AF82 E0W153 A0A182J8X5 A0A182P0K9 A0A182NV30 A0A182K3R0 A0A182LHW8 A0A2C9H6J5 A0A182RQG1 A0A084VUD8 A7UTD1 Q17PS2 A0A182WS42 A0A182U010 A0A182VL64 Q7PHM8 A0A182HXT8 D2K2F5 A0A0L0CBL7 A0A1B0A1R7 A0A224XN62 A0A1B0BQW7 A0A1A9YD57 U5ET11 A0A067R9W5 A0A336MNJ0 A0A1I8PMR0 A0A0A1WD10 T1PCA8 A0A034V7C7 A0A0K8V6P0 T1IBV5 A0A0J9RKW7 B4QD55 A0A0J9RL49 A0A0B4LGP3 Q9W0X0 Q7KVF1 W8AZM4 Q8IGY0 A0A0R3NRJ3 A0A0Q5VNZ8 B4GDA2 B4PBY8 Q292L3 B3NR92 A0A0Q5VYK1 A0A1W4V0J0 A0A0Q9WFT2 A0A3B0J111 B4MNU8 Q8SZV3 A0A3B0JMG0 B4LL12 A0A0L7LTQ3 D2A3S6 A0A0P8XQ07 B3MCD0 B4J9L8 A0A0M4EWV8 A0A1B6L2A4 A0A1B6ES61 A0A0Q9XHK8 B4KQT1 A0A1A9WI71 E9IFP2 A0A1Y1M5E8 A0A195FWH9 A0A194RIS4 A0A3Q9ELP8 A0A026WY03 A0A3L8DGW4 A0A0C9R2F4 Q24336 A0A2H1V4H1 A0A1B0GC45 A0A0J7L9G5 A0A195DD19
S4PIN9 A0A0A9Z111 A0A146L3X1 A0A0A9YZJ9 A0A1L8DJY2 A0A1L8DJU8 A0A1L8DJU1 A0A1Q3FZ01 A0A2M4CTR7 A0A1B6CLY9 A0A182QT15 A0A2M4BKE1 A0A182YBW3 A0A182WMJ8 A0A182FCS4 A0A023ETI9 A0A2M4AF82 E0W153 A0A182J8X5 A0A182P0K9 A0A182NV30 A0A182K3R0 A0A182LHW8 A0A2C9H6J5 A0A182RQG1 A0A084VUD8 A7UTD1 Q17PS2 A0A182WS42 A0A182U010 A0A182VL64 Q7PHM8 A0A182HXT8 D2K2F5 A0A0L0CBL7 A0A1B0A1R7 A0A224XN62 A0A1B0BQW7 A0A1A9YD57 U5ET11 A0A067R9W5 A0A336MNJ0 A0A1I8PMR0 A0A0A1WD10 T1PCA8 A0A034V7C7 A0A0K8V6P0 T1IBV5 A0A0J9RKW7 B4QD55 A0A0J9RL49 A0A0B4LGP3 Q9W0X0 Q7KVF1 W8AZM4 Q8IGY0 A0A0R3NRJ3 A0A0Q5VNZ8 B4GDA2 B4PBY8 Q292L3 B3NR92 A0A0Q5VYK1 A0A1W4V0J0 A0A0Q9WFT2 A0A3B0J111 B4MNU8 Q8SZV3 A0A3B0JMG0 B4LL12 A0A0L7LTQ3 D2A3S6 A0A0P8XQ07 B3MCD0 B4J9L8 A0A0M4EWV8 A0A1B6L2A4 A0A1B6ES61 A0A0Q9XHK8 B4KQT1 A0A1A9WI71 E9IFP2 A0A1Y1M5E8 A0A195FWH9 A0A194RIS4 A0A3Q9ELP8 A0A026WY03 A0A3L8DGW4 A0A0C9R2F4 Q24336 A0A2H1V4H1 A0A1B0GC45 A0A0J7L9G5 A0A195DD19
PDB
4Q4B
E-value=5.61489e-65,
Score=630
Ontologies
GO
PANTHER
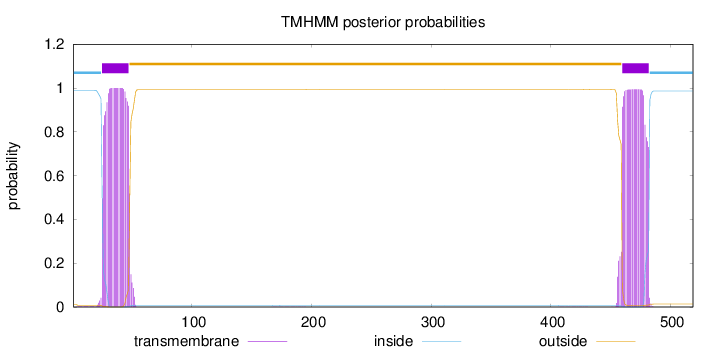
Topology
Length:
519
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.61376
Exp number, first 60 AAs:
22.86402
Total prob of N-in:
0.98857
POSSIBLE N-term signal
sequence
inside
1 - 24
TMhelix
25 - 47
outside
48 - 459
TMhelix
460 - 482
inside
483 - 519
Population Genetic Test Statistics
Pi
5.290138
Theta
4.539031
Tajima's D
0.309772
CLR
1.065109
CSRT
0.489475526223689
Interpretation
Uncertain
