Pre Gene Modal
BGIBMGA011145
Annotation
putative_fatty_acyl_reductase_FAR1_[Spodoptera_litura]
Full name
Fatty acyl-CoA reductase
Location in the cell
Cytoplasmic Reliability : 1.243 PlasmaMembrane Reliability : 1.611
Sequence
CDS
ATGGTTGCGGATATCCAAAAATATTCACCGAAGATGAAGCCAGTTGGTGAGGTGACTGGGTTCGACGCGATGGATAAGCTGGAGGCGTTGGGAGAGTCACAGATCGGGAAGATGTTCGCTGGGACCACGGTGCTACTGACCGGGGGCACAGGGTTCCTGGGGAAGCTGATGGTCGACAAACTTTTGAGATGCTGTCCGGATTTAAAAAAGCTGTTCCTACTGGCGAGACCAAAGAAAAATAAAGATATCGCGAAAAGGCTGCAGGAACAGTTTGATGACGTGCTATACGACAAGCTAAGGAAGGAGAGACCGAATTTTATAGAGAAAATATGTATAGTCGAGGGAGATGTAGGTGCTATCAACCTTGGACTCAGTCAAGAGGATCGAACGAAAATCGCTAATGAGGTTGAATTTATCTTCCACGGAGCCGCCACCGTGAGGTTCGACGAGGCTCTGAAGACAGCCGTGGAGATAAACGTGCGGGGCACCAGAGAGATGTGTATTTTGGCCAGATCCTGTGGGAAGCTGAAGGCTCTAGTTCACATTTCGACCGCATACAGTAACTGCACCGTAATGGAGATCGAAGAGAGATTCTACGAGAGCTCCCTCCCTGGTGACAAACTCATCGACCTGGTGGAAACTATGGACGAGAAGGTTATAAACAGCATCACTCCCGGTTTACTGGGGGATTTCCCGAATACGTATGCGTATACGAAATCTGTGGCCGAAGACGTGGTGCAGAAATACTCGAAGGGTCTTCCGGTGGCTTTGTTCAGGCCGTCCATAGTAATTGGAACAGCCAAAGAACCAGTCGCGGGGTGGATAGACAACGTATATGGACCTACAGGGGTTGTGGTAGGAGCCGCAGTGGGTCTACTTCACGCTCTGATCTGCAAACCCAACGCGGTAGCTGACTTGGTCCCCGGAGATATGGTTGTAAACGCTTGCATTGCAACCGCCTGGAAGACAGCCAAAGACTATCCGGGCAACCACGAGGACGCTCCACCCCCGGACCTGACGCCACCAGTTTACAACTTTGTTTCCTCAACACAGAAGCCCCTGACTTGGAGTAAATTCATGAAATACAATGAGATATATGGCCTCGAAGTGCCCACGGTGCAAGCGATTTATTACTACGTGTTTATGTTGACGTCTTCTCGTTTCGCCTACACCCTGTACTGTTTCTTCCTGCACTGGATCCCGGCCTACATCATCGATGGAGTGGCTGTGCTGATAGGCAAAAAGCCAATGCTAAGGAAAGCGTATACGAAGATAAGCAAATTCTCCGAAGTCATATCGTATTTCGCGACGCGCGAGTGGAAATTCCACAGCGGCAACACGGAGGGTCTGGTGCGGGAGCTGTGCCCGGCAGACCGGCAGACGTTCGACTTTGATTTGTCGCGGCTCGACTGGAGCGAATACTTCTACAACTACGTCAGGGGCGTCCGGGTGTACCTGCTCAAGGATCCGGTGGAAACAGTACCGGCCGGCCTCAGGAGGCATAACATACTGAAGTACTCGCACTATATCTTCTGTGCTGTATTAACGTTTGTGCTGGTCAGACTGTTGTGGGCCGCGTGGAGTCTTTTAGACCTTTAA
Protein
MVADIQKYSPKMKPVGEVTGFDAMDKLEALGESQIGKMFAGTTVLLTGGTGFLGKLMVDKLLRCCPDLKKLFLLARPKKNKDIAKRLQEQFDDVLYDKLRKERPNFIEKICIVEGDVGAINLGLSQEDRTKIANEVEFIFHGAATVRFDEALKTAVEINVRGTREMCILARSCGKLKALVHISTAYSNCTVMEIEERFYESSLPGDKLIDLVETMDEKVINSITPGLLGDFPNTYAYTKSVAEDVVQKYSKGLPVALFRPSIVIGTAKEPVAGWIDNVYGPTGVVVGAAVGLLHALICKPNAVADLVPGDMVVNACIATAWKTAKDYPGNHEDAPPPDLTPPVYNFVSSTQKPLTWSKFMKYNEIYGLEVPTVQAIYYYVFMLTSSRFAYTLYCFFLHWIPAYIIDGVAVLIGKKPMLRKAYTKISKFSEVISYFATREWKFHSGNTEGLVRELCPADRQTFDFDLSRLDWSEYFYNYVRGVRVYLLKDPVETVPAGLRRHNILKYSHYIFCAVLTFVLVRLLWAAWSLLDL
Summary
Description
Catalyzes the reduction of fatty acyl-CoA to fatty alcohols.
Catalytic Activity
a long-chain fatty acyl-CoA + 2 H(+) + 2 NADPH = a long-chain primary fatty alcohol + CoA + 2 NADP(+)
Similarity
Belongs to the fatty acyl-CoA reductase family.
Feature
chain Fatty acyl-CoA reductase
Uniprot
H9JNP1
A0A0P0EM43
A0A1V0M874
A0A437BK99
A0A2W1BH51
A0A0F6Q2N3
+ More
A0A068FK94 A0A0F6Q1J7 A0A194PTV2 A0A0N1I9W9 A0A212F0D5 A0A158NHD8 A0A151XFK4 K7J7I5 F4WU72 A0A195D4N2 K7J7I4 A0A1B6I9B4 A0A1B6IV08 A0A067QQS7 A0A1B6EVR8 A0A195EE25 A0A232EN92 A0A1B6KC16 N6TBL5 A0A1Y1ML19 A0A0M9ABB1 X1WJK9 A0A232EN78 A0A195EZP6 A0A195BRC5 A0A2S2QSR0 K7J7I3 A0A026X284 A0A195C2N5 A0A1Y1LHX9 A0A151X2I2 E9I8G2 A0A2S2QPP2 E2AWD8 A0A2S2PN15 A0A2J7QR94 A0A2J7QWL4 A0A3Q8FLY7 F4W5T8 A0A195DFI7 K7J6N1 E2BHA7 A0A0L7R054 A0A1B0CRM9 J9JN47 D2A5T7 A0A1W4W338 A0A3Q8G169 A0A2J7RNA3 J9K0H2 A0A336LZ97 A0A232F2D0 A0A1B6EEG7 E2AWD9 A0A336MV31 A0A336LCJ1 A0A2H8TQE5 A0A0T6BDN9 A0A067QUF0 A0A1B6EX35 E2AKQ3 D6WUH3 A0A1B6J450 A0A1B6HWN6 K7IT37 A0A0C9RAR7 J9JQW1 A0A2H8U048 A0A232FLM2 A0A310SC03 A0A195FYB7 A0A1B6KTN1 A0A0A9WBX3 K7J6N4 A0A1B6LVG8 A0A3Q8FT79 A0A3Q8FRZ7 A0A2J7RNB9 A0A1B6CR01 A0A1B6DLK7 D9MX52 A0A232EYC4 A0A2A3E309 V9IIS7 A0A0L7RAS9 A0A310S4M5 A0A1J1IJD8 A0A0C9PMV0 J9K312 A0A182YL41 A0A0A1X8B9
A0A068FK94 A0A0F6Q1J7 A0A194PTV2 A0A0N1I9W9 A0A212F0D5 A0A158NHD8 A0A151XFK4 K7J7I5 F4WU72 A0A195D4N2 K7J7I4 A0A1B6I9B4 A0A1B6IV08 A0A067QQS7 A0A1B6EVR8 A0A195EE25 A0A232EN92 A0A1B6KC16 N6TBL5 A0A1Y1ML19 A0A0M9ABB1 X1WJK9 A0A232EN78 A0A195EZP6 A0A195BRC5 A0A2S2QSR0 K7J7I3 A0A026X284 A0A195C2N5 A0A1Y1LHX9 A0A151X2I2 E9I8G2 A0A2S2QPP2 E2AWD8 A0A2S2PN15 A0A2J7QR94 A0A2J7QWL4 A0A3Q8FLY7 F4W5T8 A0A195DFI7 K7J6N1 E2BHA7 A0A0L7R054 A0A1B0CRM9 J9JN47 D2A5T7 A0A1W4W338 A0A3Q8G169 A0A2J7RNA3 J9K0H2 A0A336LZ97 A0A232F2D0 A0A1B6EEG7 E2AWD9 A0A336MV31 A0A336LCJ1 A0A2H8TQE5 A0A0T6BDN9 A0A067QUF0 A0A1B6EX35 E2AKQ3 D6WUH3 A0A1B6J450 A0A1B6HWN6 K7IT37 A0A0C9RAR7 J9JQW1 A0A2H8U048 A0A232FLM2 A0A310SC03 A0A195FYB7 A0A1B6KTN1 A0A0A9WBX3 K7J6N4 A0A1B6LVG8 A0A3Q8FT79 A0A3Q8FRZ7 A0A2J7RNB9 A0A1B6CR01 A0A1B6DLK7 D9MX52 A0A232EYC4 A0A2A3E309 V9IIS7 A0A0L7RAS9 A0A310S4M5 A0A1J1IJD8 A0A0C9PMV0 J9K312 A0A182YL41 A0A0A1X8B9
EC Number
1.2.1.84
Pubmed
EMBL
BABH01035163
BABH01035164
KT261695
ALJ30235.1
KU755476
ARD71186.1
+ More
RSAL01000044 RVE50678.1 KZ150217 PZC72130.1 KP237913 MF687534 AKD01766.1 ATJ44460.1 KJ622061 AID66649.1 KP237926 MF687596 AKD01779.1 ATJ44522.1 KQ459593 KPI96403.1 KQ460036 KPJ18430.1 AGBW02011118 OWR47192.1 ADTU01015701 ADTU01015702 KQ982194 KYQ59115.1 GL888353 EGI62240.1 KQ976870 KYN07801.1 GECU01024246 JAS83460.1 GECU01016929 JAS90777.1 KK853504 KDR07123.1 GECZ01027740 JAS42029.1 KQ979039 KYN23453.1 NNAY01003198 OXU19820.1 GEBQ01030984 JAT08993.1 APGK01044237 APGK01044238 KB741021 ENN75098.1 GEZM01028194 GEZM01028193 JAV86394.1 KQ435694 KOX80980.1 ABLF02017919 ABLF02017920 OXU19819.1 KQ981905 KYN33349.1 KQ976417 KYM89582.1 GGMS01011572 MBY80775.1 KK107031 QOIP01000008 EZA62096.1 RLU19323.1 KQ978379 KYM94471.1 GEZM01061226 JAV70627.1 KQ982576 KYQ54647.1 GL761538 EFZ23139.1 GGMS01010440 MBY79643.1 GL443286 EFN62238.1 GGMR01018223 MBY30842.1 NEVH01011924 PNF31087.1 NEVH01009426 PNF32977.1 MG573160 AWJ25022.1 GL887695 EGI70414.1 KQ980903 KYN11612.1 GL448287 EFN84876.1 KQ414672 KOC64230.1 AJWK01024997 AJWK01024998 AJWK01024999 ABLF02012968 KQ971345 EFA05024.1 MG573162 AWJ25024.1 NEVH01002545 PNF42320.1 ABLF02017921 UFQS01000315 UFQT01000315 SSX02758.1 SSX23130.1 NNAY01001243 OXU24628.1 GEDC01000976 JAS36322.1 EFN62239.1 UFQS01002712 UFQT01002712 SSX14582.1 SSX33980.1 SSX14581.1 SSX33979.1 GFXV01003673 MBW15478.1 LJIG01001484 KRT85456.1 KK853249 KDR09345.1 GECZ01027241 JAS42528.1 GL440368 EFN65993.1 KQ971357 EFA08474.2 GECU01013749 JAS93957.1 GECU01028612 JAS79094.1 AAZX01006711 GBYB01003931 JAG73698.1 ABLF02026798 GFXV01007576 MBW19381.1 NNAY01000061 OXU31410.1 KQ764772 OAD54398.1 KQ981208 KYN44829.1 GEBQ01025216 JAT14761.1 GBHO01038325 GBHO01038321 GBRD01012464 GDHC01011251 JAG05279.1 JAG05283.1 JAG53360.1 JAQ07378.1 GEBQ01012294 JAT27683.1 MG573171 AWJ25033.1 MG573167 AWJ25029.1 PNF42309.1 GEDC01021312 JAS15986.1 GEDC01010770 JAS26528.1 HM536636 ADI87412.1 NNAY01001641 OXU23345.1 KZ288460 PBC25481.1 JR049440 AEY60965.1 KQ414617 KOC68027.1 KQ776822 OAD52165.1 CVRI01000054 CRL00357.1 GBYB01002483 JAG72250.1 ABLF02017029 GBXI01015281 GBXI01013342 GBXI01011681 GBXI01007379 GBXI01001263 JAC99010.1 JAD00950.1 JAD02611.1 JAD06913.1 JAD13029.1
RSAL01000044 RVE50678.1 KZ150217 PZC72130.1 KP237913 MF687534 AKD01766.1 ATJ44460.1 KJ622061 AID66649.1 KP237926 MF687596 AKD01779.1 ATJ44522.1 KQ459593 KPI96403.1 KQ460036 KPJ18430.1 AGBW02011118 OWR47192.1 ADTU01015701 ADTU01015702 KQ982194 KYQ59115.1 GL888353 EGI62240.1 KQ976870 KYN07801.1 GECU01024246 JAS83460.1 GECU01016929 JAS90777.1 KK853504 KDR07123.1 GECZ01027740 JAS42029.1 KQ979039 KYN23453.1 NNAY01003198 OXU19820.1 GEBQ01030984 JAT08993.1 APGK01044237 APGK01044238 KB741021 ENN75098.1 GEZM01028194 GEZM01028193 JAV86394.1 KQ435694 KOX80980.1 ABLF02017919 ABLF02017920 OXU19819.1 KQ981905 KYN33349.1 KQ976417 KYM89582.1 GGMS01011572 MBY80775.1 KK107031 QOIP01000008 EZA62096.1 RLU19323.1 KQ978379 KYM94471.1 GEZM01061226 JAV70627.1 KQ982576 KYQ54647.1 GL761538 EFZ23139.1 GGMS01010440 MBY79643.1 GL443286 EFN62238.1 GGMR01018223 MBY30842.1 NEVH01011924 PNF31087.1 NEVH01009426 PNF32977.1 MG573160 AWJ25022.1 GL887695 EGI70414.1 KQ980903 KYN11612.1 GL448287 EFN84876.1 KQ414672 KOC64230.1 AJWK01024997 AJWK01024998 AJWK01024999 ABLF02012968 KQ971345 EFA05024.1 MG573162 AWJ25024.1 NEVH01002545 PNF42320.1 ABLF02017921 UFQS01000315 UFQT01000315 SSX02758.1 SSX23130.1 NNAY01001243 OXU24628.1 GEDC01000976 JAS36322.1 EFN62239.1 UFQS01002712 UFQT01002712 SSX14582.1 SSX33980.1 SSX14581.1 SSX33979.1 GFXV01003673 MBW15478.1 LJIG01001484 KRT85456.1 KK853249 KDR09345.1 GECZ01027241 JAS42528.1 GL440368 EFN65993.1 KQ971357 EFA08474.2 GECU01013749 JAS93957.1 GECU01028612 JAS79094.1 AAZX01006711 GBYB01003931 JAG73698.1 ABLF02026798 GFXV01007576 MBW19381.1 NNAY01000061 OXU31410.1 KQ764772 OAD54398.1 KQ981208 KYN44829.1 GEBQ01025216 JAT14761.1 GBHO01038325 GBHO01038321 GBRD01012464 GDHC01011251 JAG05279.1 JAG05283.1 JAG53360.1 JAQ07378.1 GEBQ01012294 JAT27683.1 MG573171 AWJ25033.1 MG573167 AWJ25029.1 PNF42309.1 GEDC01021312 JAS15986.1 GEDC01010770 JAS26528.1 HM536636 ADI87412.1 NNAY01001641 OXU23345.1 KZ288460 PBC25481.1 JR049440 AEY60965.1 KQ414617 KOC68027.1 KQ776822 OAD52165.1 CVRI01000054 CRL00357.1 GBYB01002483 JAG72250.1 ABLF02017029 GBXI01015281 GBXI01013342 GBXI01011681 GBXI01007379 GBXI01001263 JAC99010.1 JAD00950.1 JAD02611.1 JAD06913.1 JAD13029.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000005205
+ More
UP000075809 UP000002358 UP000007755 UP000078542 UP000027135 UP000078492 UP000215335 UP000019118 UP000053105 UP000007819 UP000078541 UP000078540 UP000053097 UP000279307 UP000000311 UP000235965 UP000008237 UP000053825 UP000092461 UP000007266 UP000192223 UP000005203 UP000242457 UP000183832 UP000076408
UP000075809 UP000002358 UP000007755 UP000078542 UP000027135 UP000078492 UP000215335 UP000019118 UP000053105 UP000007819 UP000078541 UP000078540 UP000053097 UP000279307 UP000000311 UP000235965 UP000008237 UP000053825 UP000092461 UP000007266 UP000192223 UP000005203 UP000242457 UP000183832 UP000076408
PRIDE
Interpro
SUPFAM
SSF51735
SSF51735
CDD
ProteinModelPortal
H9JNP1
A0A0P0EM43
A0A1V0M874
A0A437BK99
A0A2W1BH51
A0A0F6Q2N3
+ More
A0A068FK94 A0A0F6Q1J7 A0A194PTV2 A0A0N1I9W9 A0A212F0D5 A0A158NHD8 A0A151XFK4 K7J7I5 F4WU72 A0A195D4N2 K7J7I4 A0A1B6I9B4 A0A1B6IV08 A0A067QQS7 A0A1B6EVR8 A0A195EE25 A0A232EN92 A0A1B6KC16 N6TBL5 A0A1Y1ML19 A0A0M9ABB1 X1WJK9 A0A232EN78 A0A195EZP6 A0A195BRC5 A0A2S2QSR0 K7J7I3 A0A026X284 A0A195C2N5 A0A1Y1LHX9 A0A151X2I2 E9I8G2 A0A2S2QPP2 E2AWD8 A0A2S2PN15 A0A2J7QR94 A0A2J7QWL4 A0A3Q8FLY7 F4W5T8 A0A195DFI7 K7J6N1 E2BHA7 A0A0L7R054 A0A1B0CRM9 J9JN47 D2A5T7 A0A1W4W338 A0A3Q8G169 A0A2J7RNA3 J9K0H2 A0A336LZ97 A0A232F2D0 A0A1B6EEG7 E2AWD9 A0A336MV31 A0A336LCJ1 A0A2H8TQE5 A0A0T6BDN9 A0A067QUF0 A0A1B6EX35 E2AKQ3 D6WUH3 A0A1B6J450 A0A1B6HWN6 K7IT37 A0A0C9RAR7 J9JQW1 A0A2H8U048 A0A232FLM2 A0A310SC03 A0A195FYB7 A0A1B6KTN1 A0A0A9WBX3 K7J6N4 A0A1B6LVG8 A0A3Q8FT79 A0A3Q8FRZ7 A0A2J7RNB9 A0A1B6CR01 A0A1B6DLK7 D9MX52 A0A232EYC4 A0A2A3E309 V9IIS7 A0A0L7RAS9 A0A310S4M5 A0A1J1IJD8 A0A0C9PMV0 J9K312 A0A182YL41 A0A0A1X8B9
A0A068FK94 A0A0F6Q1J7 A0A194PTV2 A0A0N1I9W9 A0A212F0D5 A0A158NHD8 A0A151XFK4 K7J7I5 F4WU72 A0A195D4N2 K7J7I4 A0A1B6I9B4 A0A1B6IV08 A0A067QQS7 A0A1B6EVR8 A0A195EE25 A0A232EN92 A0A1B6KC16 N6TBL5 A0A1Y1ML19 A0A0M9ABB1 X1WJK9 A0A232EN78 A0A195EZP6 A0A195BRC5 A0A2S2QSR0 K7J7I3 A0A026X284 A0A195C2N5 A0A1Y1LHX9 A0A151X2I2 E9I8G2 A0A2S2QPP2 E2AWD8 A0A2S2PN15 A0A2J7QR94 A0A2J7QWL4 A0A3Q8FLY7 F4W5T8 A0A195DFI7 K7J6N1 E2BHA7 A0A0L7R054 A0A1B0CRM9 J9JN47 D2A5T7 A0A1W4W338 A0A3Q8G169 A0A2J7RNA3 J9K0H2 A0A336LZ97 A0A232F2D0 A0A1B6EEG7 E2AWD9 A0A336MV31 A0A336LCJ1 A0A2H8TQE5 A0A0T6BDN9 A0A067QUF0 A0A1B6EX35 E2AKQ3 D6WUH3 A0A1B6J450 A0A1B6HWN6 K7IT37 A0A0C9RAR7 J9JQW1 A0A2H8U048 A0A232FLM2 A0A310SC03 A0A195FYB7 A0A1B6KTN1 A0A0A9WBX3 K7J6N4 A0A1B6LVG8 A0A3Q8FT79 A0A3Q8FRZ7 A0A2J7RNB9 A0A1B6CR01 A0A1B6DLK7 D9MX52 A0A232EYC4 A0A2A3E309 V9IIS7 A0A0L7RAS9 A0A310S4M5 A0A1J1IJD8 A0A0C9PMV0 J9K312 A0A182YL41 A0A0A1X8B9
PDB
4W4T
E-value=3.84834e-09,
Score=148
Ontologies
PANTHER
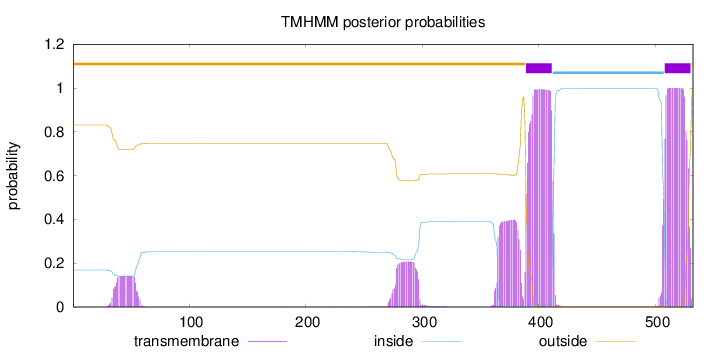
Topology
Length:
532
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
59.4463800000001
Exp number, first 60 AAs:
2.93125
Total prob of N-in:
0.16884
outside
1 - 388
TMhelix
389 - 411
inside
412 - 507
TMhelix
508 - 530
outside
531 - 532
Population Genetic Test Statistics
Pi
6.349821
Theta
13.952805
Tajima's D
-1.211505
CLR
3.539518
CSRT
0.0981950902454877
Interpretation
Uncertain
