Gene
KWMTBOMO14215 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011144
Annotation
PREDICTED:_peroxisomal_biogenesis_factor_19_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.096
Sequence
CDS
ATGTCTGACAATAAGAACGCATCCAAAAACGATCAAGATCCTGAATTAGACGATTTATTGAACAGTGCACTTCAGGATATGACAAAGAAGAAAGATGATAAGCCAAACGATACCACTCCTGATGGTGGCGAAGCAATGCAGAACATGTGGGGCGAGGAATTTATCAAAGAAGCAGCTGCTCAGTTTGAGAGCAACATGGCAGCTATCCTAGGCAGTTTCAGTGGAGTGCCAGAGGATCAGATAACACAAGAACAAATTGCATTAACTTTCAACAAAATGGCTGAAGCAGCATCACAGGTGTTAAAGCAAGGTGAATCGAGTGAAACAGAACCAAAGACAGATGACACGGCAGTACCGAAGGCAGATCAGGTCTCGGCAGCCATTTCGCAGACATTACAGAATTTGAACTCCAACTCTGAGAATCTTCAGACGCCTTTCTCAGAACAAGATCTAGCGAACATGTTTAATAATTTCAATTTAGGTGAAGGGGGCCAGGAAGGAAACATGTTCCTGCCCTTCATGCAGGGGATGATGCAGTCGCTCTTGTCGAAAGAAGTTTTATACCCATCTTTGAAGGAGCTAGTCGACAAATATCCAAGTTGGTTGAACGAGAACAAGACAAAGATTGAACAAAGCGAATACGAAAGATTCGAAAAACAGCAAACGTTAATGCAGCAAGTATGTGCTGAACTCGAGAACGAATTGGATAGTGACAGTGAAGAAGTTAAGAGAAAACGCTTCGAGAAAGTGTTGGAGTTGATGCAGAAGATGCAAGACCTAGGCCAGCCTCCCACTGAGCTGGTCGGGGATATAGGACCGGTACCTCAGAGCTTCGGTCCAGCGGCACAGGATCCATCACAATGTTGCATCAACTAG
Protein
MSDNKNASKNDQDPELDDLLNSALQDMTKKKDDKPNDTTPDGGEAMQNMWGEEFIKEAAAQFESNMAAILGSFSGVPEDQITQEQIALTFNKMAEAASQVLKQGESSETEPKTDDTAVPKADQVSAAISQTLQNLNSNSENLQTPFSEQDLANMFNNFNLGEGGQEGNMFLPFMQGMMQSLLSKEVLYPSLKELVDKYPSWLNENKTKIEQSEYERFEKQQTLMQQVCAELENELDSDSEEVKRKRFEKVLELMQKMQDLGQPPTELVGDIGPVPQSFGPAAQDPSQCCIN
Summary
Uniprot
H9JNP0
A0A3S2NWJ1
A0A2A4K2P2
A0A2W1BGR7
A0A194PZ12
A0A212F1K6
+ More
A0A0N1IHV7 A0A2H1VUK4 A0A2H1WKF7 A0A067QRF9 Q1HQH5 A0A023EQ27 A0A2J7RA55 A0A2M4AF94 A0A2M4AFH5 A0A2M3Z423 A0A2M3Z4A6 A0A2M4AFI6 W5JGH7 A0A2M4BUZ9 A0A2M4BUX1 A0A182JZM0 D2A5I0 A0A182F2U0 A0A182LSZ8 A0A182ILD8 A0A182Y9A9 A0A084VJ94 A0A182P5G8 A0A182WA04 A0A1W4WHV8 Q17BF2 A0A182QQC4 A0A182X611 A0A182UGM3 A0A182IB35 A0A182V2X6 A0A182LJZ6 A0NE60 A0A182NT58 A0A1J1IP15 A0A1Q3FBU8 B0WPW5 A0A182R2J2 A0A1Y1N8N8 A0A1B0D140 A0A182T626 A0A1L8DCX4 A0A1B0CDN0 U5ER89 K7IM63 A0A232EYX6 N6T3V9 A0A0K8TSZ6 A0A1Q3FEQ9 J3JYA1 A0A0Q9X151 C3YPM8 A0A2A3E2H6 B4N771 A0A1S3ISW8 B4P1Y4 B4IE62 B4Q3Q0 A0A1A9UL13 A0A3B0K618 Q29NK5 B3MVH8 Q8T9H4 A0A1W4VC02 E0W135 B4KI00 A0A1A9ZD26 Q8IP97 H0RND5 A0A087ZNK5 D3TMI9 A0A1A9XCR1 A0A1B0C460 A0A1I8QD73 B4JBL5 A0A0J7JZX0 E2A8G4 A0A336MIV1 T1PBP4 K1Q532 A0A0P4W0R9 B3N3X4 A0A158NN30 A0A1B0FBW1 A0A1L8EE41 A0A0N0U2J3 B4LSF4 A0A1A9WKW2 A0A0P4ZHX1
A0A0N1IHV7 A0A2H1VUK4 A0A2H1WKF7 A0A067QRF9 Q1HQH5 A0A023EQ27 A0A2J7RA55 A0A2M4AF94 A0A2M4AFH5 A0A2M3Z423 A0A2M3Z4A6 A0A2M4AFI6 W5JGH7 A0A2M4BUZ9 A0A2M4BUX1 A0A182JZM0 D2A5I0 A0A182F2U0 A0A182LSZ8 A0A182ILD8 A0A182Y9A9 A0A084VJ94 A0A182P5G8 A0A182WA04 A0A1W4WHV8 Q17BF2 A0A182QQC4 A0A182X611 A0A182UGM3 A0A182IB35 A0A182V2X6 A0A182LJZ6 A0NE60 A0A182NT58 A0A1J1IP15 A0A1Q3FBU8 B0WPW5 A0A182R2J2 A0A1Y1N8N8 A0A1B0D140 A0A182T626 A0A1L8DCX4 A0A1B0CDN0 U5ER89 K7IM63 A0A232EYX6 N6T3V9 A0A0K8TSZ6 A0A1Q3FEQ9 J3JYA1 A0A0Q9X151 C3YPM8 A0A2A3E2H6 B4N771 A0A1S3ISW8 B4P1Y4 B4IE62 B4Q3Q0 A0A1A9UL13 A0A3B0K618 Q29NK5 B3MVH8 Q8T9H4 A0A1W4VC02 E0W135 B4KI00 A0A1A9ZD26 Q8IP97 H0RND5 A0A087ZNK5 D3TMI9 A0A1A9XCR1 A0A1B0C460 A0A1I8QD73 B4JBL5 A0A0J7JZX0 E2A8G4 A0A336MIV1 T1PBP4 K1Q532 A0A0P4W0R9 B3N3X4 A0A158NN30 A0A1B0FBW1 A0A1L8EE41 A0A0N0U2J3 B4LSF4 A0A1A9WKW2 A0A0P4ZHX1
Pubmed
19121390
28756777
26354079
22118469
24845553
17204158
+ More
17510324 24945155 26483478 20920257 23761445 18362917 19820115 25244985 24438588 20966253 12364791 14747013 17210077 28004739 20075255 28648823 23537049 26369729 22516182 17994087 18563158 17550304 22936249 15632085 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 20798317 25315136 22992520 27129103 21347285
17510324 24945155 26483478 20920257 23761445 18362917 19820115 25244985 24438588 20966253 12364791 14747013 17210077 28004739 20075255 28648823 23537049 26369729 22516182 17994087 18563158 17550304 22936249 15632085 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 20798317 25315136 22992520 27129103 21347285
EMBL
BABH01035155
BABH01035156
BABH01035157
RSAL01000044
RVE50677.1
NWSH01000230
+ More
PCG78174.1 KZ150147 PZC72874.1 KQ459593 KPI96395.1 AGBW02010856 OWR47617.1 KQ460036 KPJ18437.1 ODYU01004532 SOQ44515.1 ODYU01009254 SOQ53539.1 KK853026 KDR12308.1 DQ440469 CH477322 ABF18502.1 EAT43591.1 JXUM01009874 GAPW01002949 KQ560294 JAC10649.1 KXJ83214.1 NEVH01006578 PNF37713.1 GGFK01006129 MBW39450.1 GGFK01006213 MBW39534.1 GGFM01002521 MBW23272.1 GGFM01002595 MBW23346.1 GGFK01006186 MBW39507.1 ADMH02001599 ETN61899.1 GGFJ01007738 MBW56879.1 GGFJ01007739 MBW56880.1 KQ971345 EFA05071.1 AXCM01000091 ATLV01013476 KE524860 KFB38038.1 EAT43592.1 AXCN02000729 APCN01000637 AAAB01008944 EAU76734.1 CVRI01000055 CRL01474.1 GFDL01010077 JAV24968.1 DS232030 EDS32553.1 GEZM01009822 GEZM01009821 JAV94292.1 AJVK01010182 GFDF01009920 JAV04164.1 AJWK01008028 GANO01002911 JAB56960.1 NNAY01001594 OXU23487.1 APGK01053536 KB741224 ENN72313.1 GDAI01000340 JAI17263.1 GFDL01009009 JAV26036.1 BT128225 KB631749 AEE63186.1 ERL85753.1 CH964182 KRF99251.1 GG666538 EEN57812.1 KZ288427 PBC25907.1 EDW80212.1 CM000157 EDW88155.1 CH480831 EDW45889.1 CM000361 CM002910 EDX04775.1 KMY89874.1 OUUW01000006 SPP81449.1 CH379059 EAL34214.1 CH902624 EDV33243.1 AY069294 AAL39439.1 DS235866 EEB19341.1 CH933807 EDW11282.1 AE014134 AAF53161.1 AAN10802.2 BT132857 AEV12232.1 EZ422641 ADD18917.1 JXJN01025274 CH916368 EDW04038.1 LBMM01019200 KMQ83594.1 GL437593 EFN70284.1 UFQT01001365 SSX30216.1 KA645530 AFP60159.1 JH818976 EKC26424.1 GDKW01000124 JAI56471.1 CH954177 EDV58826.1 ADTU01002487 CCAG010019860 GFDG01001956 JAV16843.1 KQ438551 KOX67202.1 CH940649 EDW64776.1 GDIP01225834 JAI97567.1
PCG78174.1 KZ150147 PZC72874.1 KQ459593 KPI96395.1 AGBW02010856 OWR47617.1 KQ460036 KPJ18437.1 ODYU01004532 SOQ44515.1 ODYU01009254 SOQ53539.1 KK853026 KDR12308.1 DQ440469 CH477322 ABF18502.1 EAT43591.1 JXUM01009874 GAPW01002949 KQ560294 JAC10649.1 KXJ83214.1 NEVH01006578 PNF37713.1 GGFK01006129 MBW39450.1 GGFK01006213 MBW39534.1 GGFM01002521 MBW23272.1 GGFM01002595 MBW23346.1 GGFK01006186 MBW39507.1 ADMH02001599 ETN61899.1 GGFJ01007738 MBW56879.1 GGFJ01007739 MBW56880.1 KQ971345 EFA05071.1 AXCM01000091 ATLV01013476 KE524860 KFB38038.1 EAT43592.1 AXCN02000729 APCN01000637 AAAB01008944 EAU76734.1 CVRI01000055 CRL01474.1 GFDL01010077 JAV24968.1 DS232030 EDS32553.1 GEZM01009822 GEZM01009821 JAV94292.1 AJVK01010182 GFDF01009920 JAV04164.1 AJWK01008028 GANO01002911 JAB56960.1 NNAY01001594 OXU23487.1 APGK01053536 KB741224 ENN72313.1 GDAI01000340 JAI17263.1 GFDL01009009 JAV26036.1 BT128225 KB631749 AEE63186.1 ERL85753.1 CH964182 KRF99251.1 GG666538 EEN57812.1 KZ288427 PBC25907.1 EDW80212.1 CM000157 EDW88155.1 CH480831 EDW45889.1 CM000361 CM002910 EDX04775.1 KMY89874.1 OUUW01000006 SPP81449.1 CH379059 EAL34214.1 CH902624 EDV33243.1 AY069294 AAL39439.1 DS235866 EEB19341.1 CH933807 EDW11282.1 AE014134 AAF53161.1 AAN10802.2 BT132857 AEV12232.1 EZ422641 ADD18917.1 JXJN01025274 CH916368 EDW04038.1 LBMM01019200 KMQ83594.1 GL437593 EFN70284.1 UFQT01001365 SSX30216.1 KA645530 AFP60159.1 JH818976 EKC26424.1 GDKW01000124 JAI56471.1 CH954177 EDV58826.1 ADTU01002487 CCAG010019860 GFDG01001956 JAV16843.1 KQ438551 KOX67202.1 CH940649 EDW64776.1 GDIP01225834 JAI97567.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000053240
+ More
UP000027135 UP000008820 UP000069940 UP000249989 UP000235965 UP000000673 UP000075881 UP000007266 UP000069272 UP000075883 UP000075880 UP000076408 UP000030765 UP000075885 UP000075920 UP000192223 UP000075886 UP000076407 UP000075902 UP000075840 UP000075903 UP000075882 UP000007062 UP000075884 UP000183832 UP000002320 UP000075900 UP000092462 UP000075901 UP000092461 UP000002358 UP000215335 UP000019118 UP000030742 UP000007798 UP000001554 UP000242457 UP000085678 UP000002282 UP000001292 UP000000304 UP000078200 UP000268350 UP000001819 UP000007801 UP000192221 UP000009046 UP000009192 UP000092445 UP000000803 UP000005203 UP000092443 UP000092460 UP000095300 UP000001070 UP000036403 UP000000311 UP000095301 UP000005408 UP000008711 UP000005205 UP000092444 UP000053105 UP000008792 UP000091820
UP000027135 UP000008820 UP000069940 UP000249989 UP000235965 UP000000673 UP000075881 UP000007266 UP000069272 UP000075883 UP000075880 UP000076408 UP000030765 UP000075885 UP000075920 UP000192223 UP000075886 UP000076407 UP000075902 UP000075840 UP000075903 UP000075882 UP000007062 UP000075884 UP000183832 UP000002320 UP000075900 UP000092462 UP000075901 UP000092461 UP000002358 UP000215335 UP000019118 UP000030742 UP000007798 UP000001554 UP000242457 UP000085678 UP000002282 UP000001292 UP000000304 UP000078200 UP000268350 UP000001819 UP000007801 UP000192221 UP000009046 UP000009192 UP000092445 UP000000803 UP000005203 UP000092443 UP000092460 UP000095300 UP000001070 UP000036403 UP000000311 UP000095301 UP000005408 UP000008711 UP000005205 UP000092444 UP000053105 UP000008792 UP000091820
Interpro
SUPFAM
SSF56784
SSF56784
Gene 3D
ProteinModelPortal
H9JNP0
A0A3S2NWJ1
A0A2A4K2P2
A0A2W1BGR7
A0A194PZ12
A0A212F1K6
+ More
A0A0N1IHV7 A0A2H1VUK4 A0A2H1WKF7 A0A067QRF9 Q1HQH5 A0A023EQ27 A0A2J7RA55 A0A2M4AF94 A0A2M4AFH5 A0A2M3Z423 A0A2M3Z4A6 A0A2M4AFI6 W5JGH7 A0A2M4BUZ9 A0A2M4BUX1 A0A182JZM0 D2A5I0 A0A182F2U0 A0A182LSZ8 A0A182ILD8 A0A182Y9A9 A0A084VJ94 A0A182P5G8 A0A182WA04 A0A1W4WHV8 Q17BF2 A0A182QQC4 A0A182X611 A0A182UGM3 A0A182IB35 A0A182V2X6 A0A182LJZ6 A0NE60 A0A182NT58 A0A1J1IP15 A0A1Q3FBU8 B0WPW5 A0A182R2J2 A0A1Y1N8N8 A0A1B0D140 A0A182T626 A0A1L8DCX4 A0A1B0CDN0 U5ER89 K7IM63 A0A232EYX6 N6T3V9 A0A0K8TSZ6 A0A1Q3FEQ9 J3JYA1 A0A0Q9X151 C3YPM8 A0A2A3E2H6 B4N771 A0A1S3ISW8 B4P1Y4 B4IE62 B4Q3Q0 A0A1A9UL13 A0A3B0K618 Q29NK5 B3MVH8 Q8T9H4 A0A1W4VC02 E0W135 B4KI00 A0A1A9ZD26 Q8IP97 H0RND5 A0A087ZNK5 D3TMI9 A0A1A9XCR1 A0A1B0C460 A0A1I8QD73 B4JBL5 A0A0J7JZX0 E2A8G4 A0A336MIV1 T1PBP4 K1Q532 A0A0P4W0R9 B3N3X4 A0A158NN30 A0A1B0FBW1 A0A1L8EE41 A0A0N0U2J3 B4LSF4 A0A1A9WKW2 A0A0P4ZHX1
A0A0N1IHV7 A0A2H1VUK4 A0A2H1WKF7 A0A067QRF9 Q1HQH5 A0A023EQ27 A0A2J7RA55 A0A2M4AF94 A0A2M4AFH5 A0A2M3Z423 A0A2M3Z4A6 A0A2M4AFI6 W5JGH7 A0A2M4BUZ9 A0A2M4BUX1 A0A182JZM0 D2A5I0 A0A182F2U0 A0A182LSZ8 A0A182ILD8 A0A182Y9A9 A0A084VJ94 A0A182P5G8 A0A182WA04 A0A1W4WHV8 Q17BF2 A0A182QQC4 A0A182X611 A0A182UGM3 A0A182IB35 A0A182V2X6 A0A182LJZ6 A0NE60 A0A182NT58 A0A1J1IP15 A0A1Q3FBU8 B0WPW5 A0A182R2J2 A0A1Y1N8N8 A0A1B0D140 A0A182T626 A0A1L8DCX4 A0A1B0CDN0 U5ER89 K7IM63 A0A232EYX6 N6T3V9 A0A0K8TSZ6 A0A1Q3FEQ9 J3JYA1 A0A0Q9X151 C3YPM8 A0A2A3E2H6 B4N771 A0A1S3ISW8 B4P1Y4 B4IE62 B4Q3Q0 A0A1A9UL13 A0A3B0K618 Q29NK5 B3MVH8 Q8T9H4 A0A1W4VC02 E0W135 B4KI00 A0A1A9ZD26 Q8IP97 H0RND5 A0A087ZNK5 D3TMI9 A0A1A9XCR1 A0A1B0C460 A0A1I8QD73 B4JBL5 A0A0J7JZX0 E2A8G4 A0A336MIV1 T1PBP4 K1Q532 A0A0P4W0R9 B3N3X4 A0A158NN30 A0A1B0FBW1 A0A1L8EE41 A0A0N0U2J3 B4LSF4 A0A1A9WKW2 A0A0P4ZHX1
PDB
5LNF
E-value=4.3269e-21,
Score=248
Ontologies
GO
PANTHER
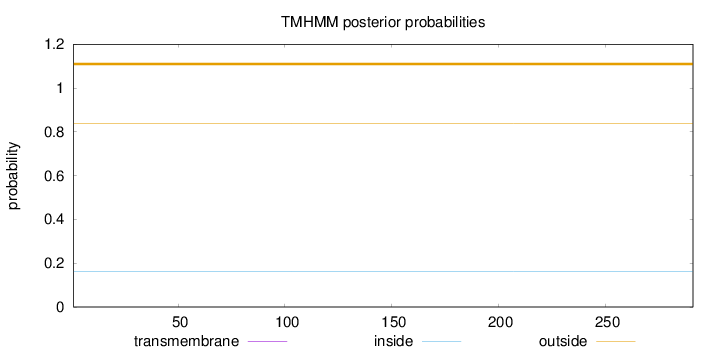
Topology
Length:
291
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00182
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.16107
outside
1 - 291
Population Genetic Test Statistics
Pi
21.227783
Theta
26.395176
Tajima's D
-0.632867
CLR
0.509322
CSRT
0.212639368031598
Interpretation
Uncertain
