Gene
KWMTBOMO14212
Pre Gene Modal
BGIBMGA011121
Annotation
PREDICTED:_fatty_acyl-CoA_reductase_1-like_isoform_X2_[Papilio_polytes]
Full name
Fatty acyl-CoA reductase
+ More
Fatty acyl-CoA reductase wat
Fatty acyl-CoA reductase wat
Alternative Name
Protein waterproof
Location in the cell
Cytoplasmic Reliability : 1.755
Sequence
CDS
ATGGTAAACACAGAGGTCCACGAAGAATTCTATCCTTGCACATGGACTCCTGAAGCTATGCTGGCGATAGCAGAGAAACTAGATGCGGAACAACTCAACGCCATCACTCCAAGTATCATAAAAGGATGGCCGAACACTTACACGTTCGAAAAAGCTTTGGCTGAAGCTTTAGTGAAGAATGCTTCGGATCTGCCCATCTGTATAGTGAGGCCCGCAGTTGTGATTGGCGCTTATTACGAACCGATGCCTGGCTGGTTGGATAAGACTTGTTTATTCGGCGCAAATGCGGTATGA
Protein
MVNTEVHEEFYPCTWTPEAMLAIAEKLDAEQLNAITPSIIKGWPNTYTFEKALAEALVKNASDLPICIVRPAVVIGAYYEPMPGWLDKTCLFGANAV
Summary
Description
Catalyzes the reduction of fatty acyl-CoA to fatty alcohols.
Catalyzes the reduction of saturated fatty acyl-CoA to fatty alcohols. The preferred substrates are C24:0 and C26:0. Necessary for the final stages of tracheal maturation, to facilitate the transition from water-filled to gas-filled tubes. May help to maintain the integrity of the outer hydrophobic envelope of the trachea.
Catalyzes the reduction of saturated fatty acyl-CoA to fatty alcohols. The preferred substrates are C24:0 and C26:0. Necessary for the final stages of tracheal maturation, to facilitate the transition from water-filled to gas-filled tubes. May help to maintain the integrity of the outer hydrophobic envelope of the trachea.
Catalytic Activity
a long-chain fatty acyl-CoA + 2 H(+) + 2 NADPH = a long-chain primary fatty alcohol + CoA + 2 NADP(+)
Similarity
Belongs to the fatty acyl-CoA reductase family.
Keywords
Cell membrane
Complete proteome
Lipid biosynthesis
Lipid metabolism
Membrane
NADP
Oxidoreductase
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Fatty acyl-CoA reductase wat
Uniprot
H9JNL7
A0A2H1VAJ7
A0A2H1WRU0
A0A2W1BRZ8
D2SNS9
A0A2H1VWV0
+ More
A0A2A4K1F7 A0A2W1BAL1 A0A0P0ELA9 A0A2A4JIF6 A0A2H1WIY5 A0A2H1WRX8 A0A2A4K1U6 A0A2H1W006 A0A291P0Q6 A0A2W1BEZ6 A0A2A4JY42 A0A2H1VVN7 A0A2W1BUR6 A0A194PUH6 A0A0N1PK47 A0A2W1B8Q0 A0A2H1WT26 A0A2A4J5Y0 A0A2A4JYT4 A0A194PV95 A0A212ER01 A0A2A4K2D3 A0A2A4JY41 A0A2H1V3I1 A0A2W1BH40 D6WUG9 A0A3B0KQF0 B3LXP4 A0A3M7QWY4 B4LXG9 B3P5N1 B4QZE6 B4N9K4 B4JRF4 A0A1W4VH99 A0A2S2R203 Q8MS59 B4PQM4 Q298Y9 B4G4W3 A0A147BN16 A0A1B6LNP5 A0A2S2PD11 D6X3Q1 B4HZ37 A0A1B6J484 T2M8I6 A0A0L0BW16 W8BGY4 A0A1I8MLW5 J9KT21 A0A1B6G8A3 A0A1B6EM21 A0A0Q9WGB4 B4M063 A0A0A9VVQ7 A0A0A9VYR3 A0A2H1VZY4 A0A0M8ZN21 A0A194PT03 E2AKA1 A0A067R2Y2 A0A2S2PW93 B4KDT7 A0A194QZH1 B5DVJ3 A0A0Q9XA67 E2BCH8 A0A194R4R4 B4LPA6 A0A1W4UAR6 A0A1W4UPL0 A0A3B0JDM3 A0A0J7KV18 E0VSV6 Q29B58 A0A0M4F301 A0A3R7QBQ9 A0A1L8DYA8 A0A1Z3FT38 D2A5A7 B4PMY1 A0A0K8W8A8 A0A2R7WZM3 B4NBE6 A0A0K8U220 A0A034W8C5 A0A2A3EL05 V9IAG7 D9MWM7 A0A195EP06 A0A069DV62 B4JTC0 A0A182G8H4
A0A2A4K1F7 A0A2W1BAL1 A0A0P0ELA9 A0A2A4JIF6 A0A2H1WIY5 A0A2H1WRX8 A0A2A4K1U6 A0A2H1W006 A0A291P0Q6 A0A2W1BEZ6 A0A2A4JY42 A0A2H1VVN7 A0A2W1BUR6 A0A194PUH6 A0A0N1PK47 A0A2W1B8Q0 A0A2H1WT26 A0A2A4J5Y0 A0A2A4JYT4 A0A194PV95 A0A212ER01 A0A2A4K2D3 A0A2A4JY41 A0A2H1V3I1 A0A2W1BH40 D6WUG9 A0A3B0KQF0 B3LXP4 A0A3M7QWY4 B4LXG9 B3P5N1 B4QZE6 B4N9K4 B4JRF4 A0A1W4VH99 A0A2S2R203 Q8MS59 B4PQM4 Q298Y9 B4G4W3 A0A147BN16 A0A1B6LNP5 A0A2S2PD11 D6X3Q1 B4HZ37 A0A1B6J484 T2M8I6 A0A0L0BW16 W8BGY4 A0A1I8MLW5 J9KT21 A0A1B6G8A3 A0A1B6EM21 A0A0Q9WGB4 B4M063 A0A0A9VVQ7 A0A0A9VYR3 A0A2H1VZY4 A0A0M8ZN21 A0A194PT03 E2AKA1 A0A067R2Y2 A0A2S2PW93 B4KDT7 A0A194QZH1 B5DVJ3 A0A0Q9XA67 E2BCH8 A0A194R4R4 B4LPA6 A0A1W4UAR6 A0A1W4UPL0 A0A3B0JDM3 A0A0J7KV18 E0VSV6 Q29B58 A0A0M4F301 A0A3R7QBQ9 A0A1L8DYA8 A0A1Z3FT38 D2A5A7 B4PMY1 A0A0K8W8A8 A0A2R7WZM3 B4NBE6 A0A0K8U220 A0A034W8C5 A0A2A3EL05 V9IAG7 D9MWM7 A0A195EP06 A0A069DV62 B4JTC0 A0A182G8H4
EC Number
1.2.1.84
Pubmed
EMBL
BABH01035147
BABH01035148
BABH01035149
ODYU01001262
SOQ37234.1
ODYU01010569
+ More
SOQ55773.1 KZ149959 PZC76395.1 EZ407212 ACX53770.1 ODYU01004617 SOQ44684.1 NWSH01000263 PCG77899.1 KZ150217 PZC72128.1 KT261699 ALJ30239.1 NWSH01001446 PCG71213.1 ODYU01008973 SOQ53018.1 ODYU01010591 SOQ55811.1 PCG77898.1 ODYU01005466 SOQ46353.1 MF687542 ATJ44468.1 KZ150147 PZC72871.1 NWSH01000368 PCG76935.1 ODYU01004416 SOQ44264.1 PZC76396.1 KQ459593 KPI96404.1 KQ460036 KPJ18429.1 PZC72129.1 ODYU01010853 SOQ56210.1 NWSH01003107 PCG66934.1 PCG76936.1 KPI96679.1 AGBW02013183 OWR43905.1 NWSH01000230 PCG78176.1 PCG76937.1 ODYU01000509 SOQ35403.1 PZC72126.1 KQ971357 EFA08470.2 OUUW01000013 SPP88066.1 CH902617 EDV43938.1 REGN01004851 RNA15897.1 CH940650 EDW67847.2 CH954182 EDV53281.1 CM000364 EDX14784.1 CH964232 EDW81680.1 CH916373 EDV94344.1 GGMS01014761 MBY83964.1 AE014297 AY119078 AAM50938.1 CM000160 EDW98363.1 CM000070 EAL27815.1 CH479179 EDW24629.1 GEGO01003258 JAR92146.1 GEBQ01014616 JAT25361.1 GGMR01014701 MBY27320.1 KQ971377 EEZ97452.1 CH480819 EDW53294.1 GECU01013738 JAS93968.1 HAAD01002192 CDG68424.1 JRES01001250 KNC24227.1 GAMC01006030 JAC00526.1 ABLF02009079 ABLF02025536 ABLF02025541 ABLF02025542 ABLF02025547 ABLF02032807 GECZ01011099 JAS58670.1 GECZ01030782 JAS38987.1 KRF83772.1 EDW68313.2 GBHO01043282 JAG00322.1 GBHO01043288 GBRD01007683 JAG00316.1 JAG58138.1 ODYU01005462 SOQ46347.1 KQ436010 KOX67575.1 KPI96442.1 GL440209 EFN66142.1 KK852740 KDR17399.1 GGMS01000518 MBY69721.1 CH933806 EDW14934.1 KQ460890 KPJ10852.1 EDY67660.1 KRG01287.1 GL447283 EFN86604.1 KPJ10851.1 CH940648 EDW60215.1 OUUW01000005 SPP80484.1 LBMM01002956 KMQ94136.1 DS235758 EEB16462.1 EAL27142.2 CP012526 ALC45532.1 QCYY01001969 ROT73914.1 GFDF01002656 JAV11428.1 KY274178 ASC44542.1 KQ971345 EFA05103.1 EDW96993.1 GDHF01004943 JAI47371.1 KK855841 PTY24075.1 EDW81110.1 GDHF01031934 JAI20380.1 GAKP01008396 GAKP01008395 JAC50557.1 KZ288220 PBC32174.1 JR038077 JR038078 AEY58108.1 AEY58109.1 HM483391 ADJ56408.1 KQ978625 KYN29624.1 GBGD01001183 JAC87706.1 EDV95010.1 JXUM01047959 KQ561540 KXJ78212.1
SOQ55773.1 KZ149959 PZC76395.1 EZ407212 ACX53770.1 ODYU01004617 SOQ44684.1 NWSH01000263 PCG77899.1 KZ150217 PZC72128.1 KT261699 ALJ30239.1 NWSH01001446 PCG71213.1 ODYU01008973 SOQ53018.1 ODYU01010591 SOQ55811.1 PCG77898.1 ODYU01005466 SOQ46353.1 MF687542 ATJ44468.1 KZ150147 PZC72871.1 NWSH01000368 PCG76935.1 ODYU01004416 SOQ44264.1 PZC76396.1 KQ459593 KPI96404.1 KQ460036 KPJ18429.1 PZC72129.1 ODYU01010853 SOQ56210.1 NWSH01003107 PCG66934.1 PCG76936.1 KPI96679.1 AGBW02013183 OWR43905.1 NWSH01000230 PCG78176.1 PCG76937.1 ODYU01000509 SOQ35403.1 PZC72126.1 KQ971357 EFA08470.2 OUUW01000013 SPP88066.1 CH902617 EDV43938.1 REGN01004851 RNA15897.1 CH940650 EDW67847.2 CH954182 EDV53281.1 CM000364 EDX14784.1 CH964232 EDW81680.1 CH916373 EDV94344.1 GGMS01014761 MBY83964.1 AE014297 AY119078 AAM50938.1 CM000160 EDW98363.1 CM000070 EAL27815.1 CH479179 EDW24629.1 GEGO01003258 JAR92146.1 GEBQ01014616 JAT25361.1 GGMR01014701 MBY27320.1 KQ971377 EEZ97452.1 CH480819 EDW53294.1 GECU01013738 JAS93968.1 HAAD01002192 CDG68424.1 JRES01001250 KNC24227.1 GAMC01006030 JAC00526.1 ABLF02009079 ABLF02025536 ABLF02025541 ABLF02025542 ABLF02025547 ABLF02032807 GECZ01011099 JAS58670.1 GECZ01030782 JAS38987.1 KRF83772.1 EDW68313.2 GBHO01043282 JAG00322.1 GBHO01043288 GBRD01007683 JAG00316.1 JAG58138.1 ODYU01005462 SOQ46347.1 KQ436010 KOX67575.1 KPI96442.1 GL440209 EFN66142.1 KK852740 KDR17399.1 GGMS01000518 MBY69721.1 CH933806 EDW14934.1 KQ460890 KPJ10852.1 EDY67660.1 KRG01287.1 GL447283 EFN86604.1 KPJ10851.1 CH940648 EDW60215.1 OUUW01000005 SPP80484.1 LBMM01002956 KMQ94136.1 DS235758 EEB16462.1 EAL27142.2 CP012526 ALC45532.1 QCYY01001969 ROT73914.1 GFDF01002656 JAV11428.1 KY274178 ASC44542.1 KQ971345 EFA05103.1 EDW96993.1 GDHF01004943 JAI47371.1 KK855841 PTY24075.1 EDW81110.1 GDHF01031934 JAI20380.1 GAKP01008396 GAKP01008395 JAC50557.1 KZ288220 PBC32174.1 JR038077 JR038078 AEY58108.1 AEY58109.1 HM483391 ADJ56408.1 KQ978625 KYN29624.1 GBGD01001183 JAC87706.1 EDV95010.1 JXUM01047959 KQ561540 KXJ78212.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000007266
+ More
UP000268350 UP000007801 UP000276133 UP000008792 UP000008711 UP000000304 UP000007798 UP000001070 UP000192221 UP000000803 UP000002282 UP000001819 UP000008744 UP000001292 UP000037069 UP000095301 UP000007819 UP000053105 UP000000311 UP000027135 UP000009192 UP000008237 UP000036403 UP000009046 UP000092553 UP000283509 UP000242457 UP000005203 UP000078492 UP000069940 UP000249989
UP000268350 UP000007801 UP000276133 UP000008792 UP000008711 UP000000304 UP000007798 UP000001070 UP000192221 UP000000803 UP000002282 UP000001819 UP000008744 UP000001292 UP000037069 UP000095301 UP000007819 UP000053105 UP000000311 UP000027135 UP000009192 UP000008237 UP000036403 UP000009046 UP000092553 UP000283509 UP000242457 UP000005203 UP000078492 UP000069940 UP000249989
Interpro
CDD
ProteinModelPortal
H9JNL7
A0A2H1VAJ7
A0A2H1WRU0
A0A2W1BRZ8
D2SNS9
A0A2H1VWV0
+ More
A0A2A4K1F7 A0A2W1BAL1 A0A0P0ELA9 A0A2A4JIF6 A0A2H1WIY5 A0A2H1WRX8 A0A2A4K1U6 A0A2H1W006 A0A291P0Q6 A0A2W1BEZ6 A0A2A4JY42 A0A2H1VVN7 A0A2W1BUR6 A0A194PUH6 A0A0N1PK47 A0A2W1B8Q0 A0A2H1WT26 A0A2A4J5Y0 A0A2A4JYT4 A0A194PV95 A0A212ER01 A0A2A4K2D3 A0A2A4JY41 A0A2H1V3I1 A0A2W1BH40 D6WUG9 A0A3B0KQF0 B3LXP4 A0A3M7QWY4 B4LXG9 B3P5N1 B4QZE6 B4N9K4 B4JRF4 A0A1W4VH99 A0A2S2R203 Q8MS59 B4PQM4 Q298Y9 B4G4W3 A0A147BN16 A0A1B6LNP5 A0A2S2PD11 D6X3Q1 B4HZ37 A0A1B6J484 T2M8I6 A0A0L0BW16 W8BGY4 A0A1I8MLW5 J9KT21 A0A1B6G8A3 A0A1B6EM21 A0A0Q9WGB4 B4M063 A0A0A9VVQ7 A0A0A9VYR3 A0A2H1VZY4 A0A0M8ZN21 A0A194PT03 E2AKA1 A0A067R2Y2 A0A2S2PW93 B4KDT7 A0A194QZH1 B5DVJ3 A0A0Q9XA67 E2BCH8 A0A194R4R4 B4LPA6 A0A1W4UAR6 A0A1W4UPL0 A0A3B0JDM3 A0A0J7KV18 E0VSV6 Q29B58 A0A0M4F301 A0A3R7QBQ9 A0A1L8DYA8 A0A1Z3FT38 D2A5A7 B4PMY1 A0A0K8W8A8 A0A2R7WZM3 B4NBE6 A0A0K8U220 A0A034W8C5 A0A2A3EL05 V9IAG7 D9MWM7 A0A195EP06 A0A069DV62 B4JTC0 A0A182G8H4
A0A2A4K1F7 A0A2W1BAL1 A0A0P0ELA9 A0A2A4JIF6 A0A2H1WIY5 A0A2H1WRX8 A0A2A4K1U6 A0A2H1W006 A0A291P0Q6 A0A2W1BEZ6 A0A2A4JY42 A0A2H1VVN7 A0A2W1BUR6 A0A194PUH6 A0A0N1PK47 A0A2W1B8Q0 A0A2H1WT26 A0A2A4J5Y0 A0A2A4JYT4 A0A194PV95 A0A212ER01 A0A2A4K2D3 A0A2A4JY41 A0A2H1V3I1 A0A2W1BH40 D6WUG9 A0A3B0KQF0 B3LXP4 A0A3M7QWY4 B4LXG9 B3P5N1 B4QZE6 B4N9K4 B4JRF4 A0A1W4VH99 A0A2S2R203 Q8MS59 B4PQM4 Q298Y9 B4G4W3 A0A147BN16 A0A1B6LNP5 A0A2S2PD11 D6X3Q1 B4HZ37 A0A1B6J484 T2M8I6 A0A0L0BW16 W8BGY4 A0A1I8MLW5 J9KT21 A0A1B6G8A3 A0A1B6EM21 A0A0Q9WGB4 B4M063 A0A0A9VVQ7 A0A0A9VYR3 A0A2H1VZY4 A0A0M8ZN21 A0A194PT03 E2AKA1 A0A067R2Y2 A0A2S2PW93 B4KDT7 A0A194QZH1 B5DVJ3 A0A0Q9XA67 E2BCH8 A0A194R4R4 B4LPA6 A0A1W4UAR6 A0A1W4UPL0 A0A3B0JDM3 A0A0J7KV18 E0VSV6 Q29B58 A0A0M4F301 A0A3R7QBQ9 A0A1L8DYA8 A0A1Z3FT38 D2A5A7 B4PMY1 A0A0K8W8A8 A0A2R7WZM3 B4NBE6 A0A0K8U220 A0A034W8C5 A0A2A3EL05 V9IAG7 D9MWM7 A0A195EP06 A0A069DV62 B4JTC0 A0A182G8H4
Ontologies
GO
PANTHER
Topology
Subcellular location
Apical cell membrane
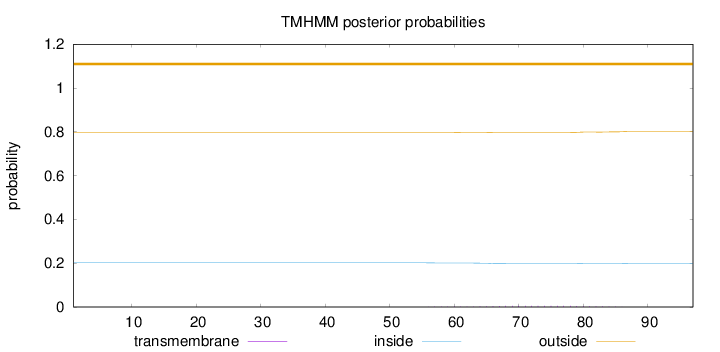
Length:
97
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10147
Exp number, first 60 AAs:
0.0067
Total prob of N-in:
0.20237
outside
1 - 97
Population Genetic Test Statistics
Pi
21.42643
Theta
16.629205
Tajima's D
0.838395
CLR
1.009546
CSRT
0.621618919054047
Interpretation
Uncertain
