Gene
KWMTBOMO14209
Pre Gene Modal
BGIBMGA011123
Annotation
hypothetical_protein_KGM_13923_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.92 Mitochondrial Reliability : 1.58
Sequence
CDS
ATGAGCGTTACCGATCCAGAGGTTAAGCACGGTAAGCCAAAGCCGGACATTTATTTGGTTGCCGCTTCGAGATTTCCCGACAAGCCGAAACCAATTAAATGCCTGGTGTTTGAGGATTCGGTGGTCGGTGTGGAGGCCGCCATCAAAGCTGGAATGCAGGTGGTCATGACCCCTGGACCTCGTCTGCAGCGAGAGCACACCCGTCACGCCACGCTCGTCATCAAGTCACTCCTGGACTTCAAACCCGAACTCTTCGGTTTGCCGCCCTTTGACAAGACTTTGAATAAAGCAACAGAAATAACCAATAAATGA
Protein
MSVTDPEVKHGKPKPDIYLVAASRFPDKPKPIKCLVFEDSVVGVEAAIKAGMQVVMTPGPRLQREHTRHATLVIKSLLDFKPELFGLPPFDKTLNKATEITNK
Summary
Uniprot
A0A212F018
A0A2W1BX41
A0A2A4K3D9
A0A3S2N7X8
A0A3R7NW74
A0A2W1BMG2
+ More
A0A2H1VHL6 A0A2H1WMW9 A0A2P2I4A9 A0A212F8Q0 A0A1A9ZCS0 A0A2P8XJF3 A0A2A4JAX9 A0A194QZP8 A0A2M4C050 A0A1A9XJN8 A0A2H1VEM3 A0A0M4EEI1 A0A2M4AP32 A0A1A9WKN8 A0A0Q9X5P5 A0A2M4BY45 T1DSD2 S4PQ87 A0A1A9VW30 A0A2M4CMY4 A0A2M4CNZ9 B4KEQ9 A0A1B0C303 A0A2W1BQK6 W5JCB8 A0A2M4ALP7 A0T4G2 A0A2M4BXX4 A0A2M3Z4S4 A0A182FR97 B4LT64 A0A194Q1D5 S4PLG4 A0A182HCU5 A0A0Q9WNL2 A0A182RDE2 A0A194PJM2 H9JL36 A0A1W7RGW9 F5HLS5 W8BI78 A0A2M4ANA3 A0A2M4CTV4 A0A182XKA5 A0A2Y9D0Y1 W8C406 A0A1B6I4X0 A0A3B0JHT9 H9JL37 A0A1B6KEN4 A0A182NSU3 A0A2H1WL50 A0A194PSI1 A0A194Q209 A0A0R3NZ21 A0A3B0K4J0 A0A2A4JLP6 A0A182K1W3 A0A182JN88 A0A2W1BQT3 A0A182PCV0 Q29LK4 A0A195CY64 A0A195AVH3 A0A158P248 A0A151XCE6 A0A2A4J9F2 A0A151IEQ1 A0A182QUH8 A0A1B6C8D4 A0A224Z5C0 A0A195E6Q3 A0A194Q1A8 B4GPT8 A0A154P1Q9 L7M167 A0A182M1D9 A0A182Y4L1 A0A1B6G3U6 A0A182TPL4 A0A194QYZ7 A0A182S823 A0A212FKC6 A0A2W1BVK3 A0A182VFD8 V5G133 A0A1E1X0U5 E9IV40 B4MTX6 A0A131Z0D8 A0A0P5BZX5 E2BE28 A0A151NWK4
A0A2H1VHL6 A0A2H1WMW9 A0A2P2I4A9 A0A212F8Q0 A0A1A9ZCS0 A0A2P8XJF3 A0A2A4JAX9 A0A194QZP8 A0A2M4C050 A0A1A9XJN8 A0A2H1VEM3 A0A0M4EEI1 A0A2M4AP32 A0A1A9WKN8 A0A0Q9X5P5 A0A2M4BY45 T1DSD2 S4PQ87 A0A1A9VW30 A0A2M4CMY4 A0A2M4CNZ9 B4KEQ9 A0A1B0C303 A0A2W1BQK6 W5JCB8 A0A2M4ALP7 A0T4G2 A0A2M4BXX4 A0A2M3Z4S4 A0A182FR97 B4LT64 A0A194Q1D5 S4PLG4 A0A182HCU5 A0A0Q9WNL2 A0A182RDE2 A0A194PJM2 H9JL36 A0A1W7RGW9 F5HLS5 W8BI78 A0A2M4ANA3 A0A2M4CTV4 A0A182XKA5 A0A2Y9D0Y1 W8C406 A0A1B6I4X0 A0A3B0JHT9 H9JL37 A0A1B6KEN4 A0A182NSU3 A0A2H1WL50 A0A194PSI1 A0A194Q209 A0A0R3NZ21 A0A3B0K4J0 A0A2A4JLP6 A0A182K1W3 A0A182JN88 A0A2W1BQT3 A0A182PCV0 Q29LK4 A0A195CY64 A0A195AVH3 A0A158P248 A0A151XCE6 A0A2A4J9F2 A0A151IEQ1 A0A182QUH8 A0A1B6C8D4 A0A224Z5C0 A0A195E6Q3 A0A194Q1A8 B4GPT8 A0A154P1Q9 L7M167 A0A182M1D9 A0A182Y4L1 A0A1B6G3U6 A0A182TPL4 A0A194QYZ7 A0A182S823 A0A212FKC6 A0A2W1BVK3 A0A182VFD8 V5G133 A0A1E1X0U5 E9IV40 B4MTX6 A0A131Z0D8 A0A0P5BZX5 E2BE28 A0A151NWK4
Pubmed
EMBL
AGBW02011196
OWR47072.1
KZ149930
PZC77380.1
NWSH01000230
PCG78172.1
+ More
RSAL01001986 RVE40613.1 QCYY01002667 ROT68424.1 KZ150147 PZC72873.1 ODYU01002607 SOQ40330.1 ODYU01009402 SOQ53804.1 IACF01003097 LAB68726.1 AGBW02009685 OWR50114.1 PYGN01001923 PSN32125.1 NWSH01002306 PCG68580.1 KQ460930 KPJ10759.1 GGFJ01009370 MBW58511.1 ODYU01002150 SOQ39275.1 CP012523 ALC38555.1 GGFK01009225 MBW42546.1 CH933807 KRG03519.1 GGFJ01008792 MBW57933.1 GAMD01001554 JAB00037.1 GAIX01001895 JAA90665.1 GGFL01002461 MBW66639.1 GGFL01002460 MBW66638.1 EDW12959.2 JXJN01024742 PZC77379.1 ADMH02001839 ETN60968.1 GGFK01008394 MBW41715.1 AAAB01008904 EAA44497.5 GGFJ01008794 MBW57935.1 GGFM01002768 MBW23519.1 CH940649 EDW64906.2 KRF81935.1 KQ459579 KPI99366.1 GAIX01004040 JAA88520.1 JXUM01128127 JXUM01128128 KQ567279 KXJ69498.1 KRF81936.1 KQ459602 KPI93626.1 BABH01011545 GDAY02001035 JAV50392.1 EGK97236.1 GAMC01009877 JAB96678.1 GGFK01008767 MBW42088.1 GGFL01004589 MBW68767.1 APCN01002192 GAMC01009876 JAB96679.1 GECU01025761 JAS81945.1 OUUW01000004 SPP79872.1 GEBQ01030092 JAT09885.1 SOQ53803.1 KQ459593 KPI96396.1 KPI99368.1 CH379060 KRT04420.1 KRT04421.1 SPP79871.1 NWSH01001155 PCG72330.1 PZC77378.1 EAL34041.3 KRT04422.1 KQ977115 KYN05610.1 KQ976731 KYM76233.1 ADTU01006978 KQ982314 KYQ58023.1 PCG68581.1 KQ977858 KYM99286.1 AXCN02000301 GEDC01027540 GEDC01019487 JAS09758.1 JAS17811.1 GFPF01011115 MAA22261.1 KQ979568 KYN20771.1 KPI99367.1 CH479187 EDW39610.1 KQ434786 KZC05168.1 GACK01008146 JAA56888.1 AXCM01002171 GECZ01012659 JAS57110.1 KPJ10758.1 AGBW02008070 OWR54191.1 KZ149923 PZC77675.1 GALX01004746 JAB63720.1 GFAC01006311 JAT92877.1 GL766116 EFZ15560.1 CH963852 EDW75565.2 GEDV01004362 JAP84195.1 GDIP01182300 JAJ41102.1 GL447755 EFN86015.1 AKHW03001669 KYO41113.1
RSAL01001986 RVE40613.1 QCYY01002667 ROT68424.1 KZ150147 PZC72873.1 ODYU01002607 SOQ40330.1 ODYU01009402 SOQ53804.1 IACF01003097 LAB68726.1 AGBW02009685 OWR50114.1 PYGN01001923 PSN32125.1 NWSH01002306 PCG68580.1 KQ460930 KPJ10759.1 GGFJ01009370 MBW58511.1 ODYU01002150 SOQ39275.1 CP012523 ALC38555.1 GGFK01009225 MBW42546.1 CH933807 KRG03519.1 GGFJ01008792 MBW57933.1 GAMD01001554 JAB00037.1 GAIX01001895 JAA90665.1 GGFL01002461 MBW66639.1 GGFL01002460 MBW66638.1 EDW12959.2 JXJN01024742 PZC77379.1 ADMH02001839 ETN60968.1 GGFK01008394 MBW41715.1 AAAB01008904 EAA44497.5 GGFJ01008794 MBW57935.1 GGFM01002768 MBW23519.1 CH940649 EDW64906.2 KRF81935.1 KQ459579 KPI99366.1 GAIX01004040 JAA88520.1 JXUM01128127 JXUM01128128 KQ567279 KXJ69498.1 KRF81936.1 KQ459602 KPI93626.1 BABH01011545 GDAY02001035 JAV50392.1 EGK97236.1 GAMC01009877 JAB96678.1 GGFK01008767 MBW42088.1 GGFL01004589 MBW68767.1 APCN01002192 GAMC01009876 JAB96679.1 GECU01025761 JAS81945.1 OUUW01000004 SPP79872.1 GEBQ01030092 JAT09885.1 SOQ53803.1 KQ459593 KPI96396.1 KPI99368.1 CH379060 KRT04420.1 KRT04421.1 SPP79871.1 NWSH01001155 PCG72330.1 PZC77378.1 EAL34041.3 KRT04422.1 KQ977115 KYN05610.1 KQ976731 KYM76233.1 ADTU01006978 KQ982314 KYQ58023.1 PCG68581.1 KQ977858 KYM99286.1 AXCN02000301 GEDC01027540 GEDC01019487 JAS09758.1 JAS17811.1 GFPF01011115 MAA22261.1 KQ979568 KYN20771.1 KPI99367.1 CH479187 EDW39610.1 KQ434786 KZC05168.1 GACK01008146 JAA56888.1 AXCM01002171 GECZ01012659 JAS57110.1 KPJ10758.1 AGBW02008070 OWR54191.1 KZ149923 PZC77675.1 GALX01004746 JAB63720.1 GFAC01006311 JAT92877.1 GL766116 EFZ15560.1 CH963852 EDW75565.2 GEDV01004362 JAP84195.1 GDIP01182300 JAJ41102.1 GL447755 EFN86015.1 AKHW03001669 KYO41113.1
Proteomes
UP000007151
UP000218220
UP000283053
UP000283509
UP000092445
UP000245037
+ More
UP000053240 UP000092443 UP000092553 UP000091820 UP000009192 UP000078200 UP000092460 UP000000673 UP000007062 UP000069272 UP000008792 UP000053268 UP000069940 UP000249989 UP000075900 UP000005204 UP000076407 UP000075840 UP000268350 UP000075884 UP000001819 UP000075881 UP000075880 UP000075885 UP000078542 UP000078540 UP000005205 UP000075809 UP000075886 UP000078492 UP000008744 UP000076502 UP000075883 UP000076408 UP000075902 UP000075901 UP000075903 UP000007798 UP000008237 UP000050525
UP000053240 UP000092443 UP000092553 UP000091820 UP000009192 UP000078200 UP000092460 UP000000673 UP000007062 UP000069272 UP000008792 UP000053268 UP000069940 UP000249989 UP000075900 UP000005204 UP000076407 UP000075840 UP000268350 UP000075884 UP000001819 UP000075881 UP000075880 UP000075885 UP000078542 UP000078540 UP000005205 UP000075809 UP000075886 UP000078492 UP000008744 UP000076502 UP000075883 UP000076408 UP000075902 UP000075901 UP000075903 UP000007798 UP000008237 UP000050525
Pfam
PF13419 HAD_2
Interpro
SUPFAM
SSF56784
SSF56784
Gene 3D
ProteinModelPortal
A0A212F018
A0A2W1BX41
A0A2A4K3D9
A0A3S2N7X8
A0A3R7NW74
A0A2W1BMG2
+ More
A0A2H1VHL6 A0A2H1WMW9 A0A2P2I4A9 A0A212F8Q0 A0A1A9ZCS0 A0A2P8XJF3 A0A2A4JAX9 A0A194QZP8 A0A2M4C050 A0A1A9XJN8 A0A2H1VEM3 A0A0M4EEI1 A0A2M4AP32 A0A1A9WKN8 A0A0Q9X5P5 A0A2M4BY45 T1DSD2 S4PQ87 A0A1A9VW30 A0A2M4CMY4 A0A2M4CNZ9 B4KEQ9 A0A1B0C303 A0A2W1BQK6 W5JCB8 A0A2M4ALP7 A0T4G2 A0A2M4BXX4 A0A2M3Z4S4 A0A182FR97 B4LT64 A0A194Q1D5 S4PLG4 A0A182HCU5 A0A0Q9WNL2 A0A182RDE2 A0A194PJM2 H9JL36 A0A1W7RGW9 F5HLS5 W8BI78 A0A2M4ANA3 A0A2M4CTV4 A0A182XKA5 A0A2Y9D0Y1 W8C406 A0A1B6I4X0 A0A3B0JHT9 H9JL37 A0A1B6KEN4 A0A182NSU3 A0A2H1WL50 A0A194PSI1 A0A194Q209 A0A0R3NZ21 A0A3B0K4J0 A0A2A4JLP6 A0A182K1W3 A0A182JN88 A0A2W1BQT3 A0A182PCV0 Q29LK4 A0A195CY64 A0A195AVH3 A0A158P248 A0A151XCE6 A0A2A4J9F2 A0A151IEQ1 A0A182QUH8 A0A1B6C8D4 A0A224Z5C0 A0A195E6Q3 A0A194Q1A8 B4GPT8 A0A154P1Q9 L7M167 A0A182M1D9 A0A182Y4L1 A0A1B6G3U6 A0A182TPL4 A0A194QYZ7 A0A182S823 A0A212FKC6 A0A2W1BVK3 A0A182VFD8 V5G133 A0A1E1X0U5 E9IV40 B4MTX6 A0A131Z0D8 A0A0P5BZX5 E2BE28 A0A151NWK4
A0A2H1VHL6 A0A2H1WMW9 A0A2P2I4A9 A0A212F8Q0 A0A1A9ZCS0 A0A2P8XJF3 A0A2A4JAX9 A0A194QZP8 A0A2M4C050 A0A1A9XJN8 A0A2H1VEM3 A0A0M4EEI1 A0A2M4AP32 A0A1A9WKN8 A0A0Q9X5P5 A0A2M4BY45 T1DSD2 S4PQ87 A0A1A9VW30 A0A2M4CMY4 A0A2M4CNZ9 B4KEQ9 A0A1B0C303 A0A2W1BQK6 W5JCB8 A0A2M4ALP7 A0T4G2 A0A2M4BXX4 A0A2M3Z4S4 A0A182FR97 B4LT64 A0A194Q1D5 S4PLG4 A0A182HCU5 A0A0Q9WNL2 A0A182RDE2 A0A194PJM2 H9JL36 A0A1W7RGW9 F5HLS5 W8BI78 A0A2M4ANA3 A0A2M4CTV4 A0A182XKA5 A0A2Y9D0Y1 W8C406 A0A1B6I4X0 A0A3B0JHT9 H9JL37 A0A1B6KEN4 A0A182NSU3 A0A2H1WL50 A0A194PSI1 A0A194Q209 A0A0R3NZ21 A0A3B0K4J0 A0A2A4JLP6 A0A182K1W3 A0A182JN88 A0A2W1BQT3 A0A182PCV0 Q29LK4 A0A195CY64 A0A195AVH3 A0A158P248 A0A151XCE6 A0A2A4J9F2 A0A151IEQ1 A0A182QUH8 A0A1B6C8D4 A0A224Z5C0 A0A195E6Q3 A0A194Q1A8 B4GPT8 A0A154P1Q9 L7M167 A0A182M1D9 A0A182Y4L1 A0A1B6G3U6 A0A182TPL4 A0A194QYZ7 A0A182S823 A0A212FKC6 A0A2W1BVK3 A0A182VFD8 V5G133 A0A1E1X0U5 E9IV40 B4MTX6 A0A131Z0D8 A0A0P5BZX5 E2BE28 A0A151NWK4
PDB
3L5K
E-value=1.62448e-23,
Score=264
Ontologies
GO
Topology
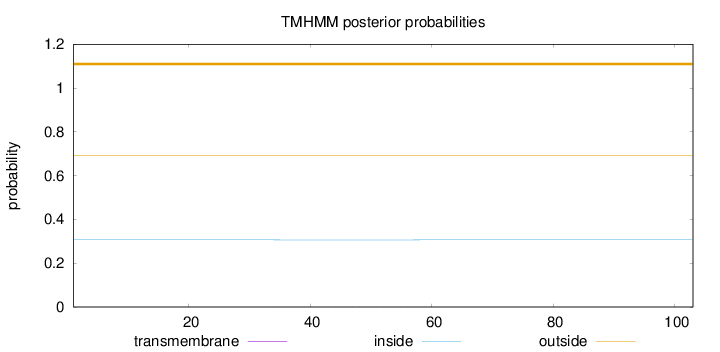
Length:
103
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01249
Exp number, first 60 AAs:
0.01173
Total prob of N-in:
0.30672
outside
1 - 103
Population Genetic Test Statistics
Pi
17.763321
Theta
15.685162
Tajima's D
-0.292056
CLR
0.804122
CSRT
0.301734913254337
Interpretation
Uncertain
