Gene
KWMTBOMO14206
Pre Gene Modal
BGIBMGA009311
Annotation
PREDICTED:_uncharacterized_protein_LOC105842116_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.392
Sequence
CDS
ATGGATTCGTTAAAACCACCGCCACCATTATGCCTCATGGGCGACTTGAAACAAAATTTTCAAAAATGGACACAGCAATATGATATTTATATGATTGCTAGCGGGACGGAAGGTGATGAAAAAATTAAAGATAAGCAGAAAAAGGCAATCTATTTGCACTGCCTAGGTGCAGAAGCACTAACCATATACAATGGTTTTAAGAACAAAGAAAAGTTATCTTATGTGGAAGTAAAACAGAAGTTTAATGAATATTGTTCGCCACCCGGAAATGAAACTTTTAGCAGACATGTCTTCTTCAGCAGAAAAAAGAGAGATGGAGAAAGTTTTGATGAATTTTTCTGTACGTTGAGAAAATTGGCGGGAGACTGTGGGTTTGCAGACCTAGAAGAAAGCTTAATAAGAGACCAAATCATATTAGGCTTAAATGATGCCAAATTAACAGAAAGATTGCTTAAGGAATCGACCTTAACATTGGATAGCTGCATGAAAATATGTAAAGTGGCTAAGAGTGCTGCAAAACAGGTTCAAGTCATGACAGAAAAGACAGTTGAAGCTGTTACAAGAAAACAAGAATCTCGAAGCAGAATGCCTAATCAGAATAAAACCAGTTTTAATAATAAACAACATCTAGGTAAGAGTGCAGAAGGATCAGTAAGGTTGAAGAATAATAATTGTGAAAGATGCGGAGGAGTGCATTCTCATCGATCCTGCCCAGCCTTTGGAAGGAGATGTGCTAAGTGTAATCGATATGGACATTACATGCAGATGTGCAGAACAGTTGGTGCTGTGTATGTCAGCGATGATGATAGTGATGACTCATTAGTAGTGGGGTCAGTAACTGTGGGCCAGGTTGATACAGAGGACTGGATCGTTGAGGCAAAAGTGAGAGGACAGAAGATGAAGTTCAAAGTGGATACAGGATCGCAGGTTAATATTTTGACTAGTGCTATAGTGAAAAAACTAGACTTAAAAATGACAGAAACTTGTACAAAATTAAAAAACTTTGATGGCTCAACTATAGAAACAGTAGGTATGACTAAAACTATAGTGAAATTCAATAAAAAAAAGTATGTACTTGACTTTTATGTGGTAAAATTTAAAACTACTAATGTGCTGAGCTATAAAGCTGCTGTGCAGTTGGGTTTAATAAACAAAGTTGAAAATATTTCAAAGGAATATGTAGTTCAGGACATTGGTGAATACAAAGATTTGTTTCATGGCTTAGGAAAATTGAACTATGTATACAAAATTAAATTAACAGACAATTGCTAA
Protein
MDSLKPPPPLCLMGDLKQNFQKWTQQYDIYMIASGTEGDEKIKDKQKKAIYLHCLGAEALTIYNGFKNKEKLSYVEVKQKFNEYCSPPGNETFSRHVFFSRKKRDGESFDEFFCTLRKLAGDCGFADLEESLIRDQIILGLNDAKLTERLLKESTLTLDSCMKICKVAKSAAKQVQVMTEKTVEAVTRKQESRSRMPNQNKTSFNNKQHLGKSAEGSVRLKNNNCERCGGVHSHRSCPAFGRRCAKCNRYGHYMQMCRTVGAVYVSDDDSDDSLVVGSVTVGQVDTEDWIVEAKVRGQKMKFKVDTGSQVNILTSAIVKKLDLKMTETCTKLKNFDGSTIETVGMTKTIVKFNKKKYVLDFYVVKFKTTNVLSYKAAVQLGLINKVENISKEYVVQDIGEYKDLFHGLGKLNYVYKIKLTDNC
Summary
Uniprot
A0A087U9K1
A0A2G8KKG5
A0A1Y1KPK9
A0A3P8Q6C6
A0A3P9HT22
A0A3B3I1S4
+ More
A0A3B3C9X5 A0A3P8S414 A0A1A8UGA0 A0A1A8D4P4 A0A3P8RHF8 A0A3B1JKR1 A0A3P8RKT7 X1WTY2 A0A3P9M8I3 I3KX76 I3KYE2 A0A146TAZ6 A0A3P9AYU2 A0A2S2N8I3 A0A1A8IVJ0 A0A3P8QAD1 A0A3P8PD24 A0A3B3HLX7 A0A2G8LMB1 A0A3P9HXP1 L7LWW6 A0A2G8L9H5 A0A146SN99 A0A1Y1MUX5 A0A1Y1LNL5 A0A0A9WVK6 A0A1Y1LVD1 A0A212FHD4 A0A3Q0J4U3 A0A2G8KBJ0 A0A2G8KFU3 A0A1E1XPH1 J9LDY4 A0A2S2NI46 A0A1Y1JV53 A0A147BKR8 J9LYD2 A0A1Y1K454 A0A1Y1M671 A0A2G8KHQ8 J9M4C2 A0A1Y1NHV7 A0A0P4VZQ4 J9M6I9 A0A0A9WHB1 A0A147BBY0 J9LNB8 A0A0A9Z4Y4 A0A2G8KB94 A0A087UXZ8 A0A224Z9B6 A0A146LC45 A0A3Q2XZ76 A0A1Y1MZH6 A0A1B8XWD8 A0A1Y1L7V1 J9KCS2 A0A1S3DM59 A0A2S2PCF5 A0A1Y1LR12 X1XQZ7 A0A0A9XW98 A0A023EXW7 A0A1Y1LZ99 A0A1A8JBP0 A0A023EWW4 A0A1S3JEC7 A0A075EIL5 A0A147BIX9 J9L288 A0A1Y1NN34 A0A293LIZ7 J9JQ26 A0A3S3NA82 A0A131Y4S4 A0A2G8JJY8 A0A2I4C5Y1 A0A3S2N4F0 A0A2B4S589 A0A0A9YK20 A0A1S3ICH8 A0A069DZS8 J9L8Q1 A0A3P9CYY5
A0A3B3C9X5 A0A3P8S414 A0A1A8UGA0 A0A1A8D4P4 A0A3P8RHF8 A0A3B1JKR1 A0A3P8RKT7 X1WTY2 A0A3P9M8I3 I3KX76 I3KYE2 A0A146TAZ6 A0A3P9AYU2 A0A2S2N8I3 A0A1A8IVJ0 A0A3P8QAD1 A0A3P8PD24 A0A3B3HLX7 A0A2G8LMB1 A0A3P9HXP1 L7LWW6 A0A2G8L9H5 A0A146SN99 A0A1Y1MUX5 A0A1Y1LNL5 A0A0A9WVK6 A0A1Y1LVD1 A0A212FHD4 A0A3Q0J4U3 A0A2G8KBJ0 A0A2G8KFU3 A0A1E1XPH1 J9LDY4 A0A2S2NI46 A0A1Y1JV53 A0A147BKR8 J9LYD2 A0A1Y1K454 A0A1Y1M671 A0A2G8KHQ8 J9M4C2 A0A1Y1NHV7 A0A0P4VZQ4 J9M6I9 A0A0A9WHB1 A0A147BBY0 J9LNB8 A0A0A9Z4Y4 A0A2G8KB94 A0A087UXZ8 A0A224Z9B6 A0A146LC45 A0A3Q2XZ76 A0A1Y1MZH6 A0A1B8XWD8 A0A1Y1L7V1 J9KCS2 A0A1S3DM59 A0A2S2PCF5 A0A1Y1LR12 X1XQZ7 A0A0A9XW98 A0A023EXW7 A0A1Y1LZ99 A0A1A8JBP0 A0A023EWW4 A0A1S3JEC7 A0A075EIL5 A0A147BIX9 J9L288 A0A1Y1NN34 A0A293LIZ7 J9JQ26 A0A3S3NA82 A0A131Y4S4 A0A2G8JJY8 A0A2I4C5Y1 A0A3S2N4F0 A0A2B4S589 A0A0A9YK20 A0A1S3ICH8 A0A069DZS8 J9L8Q1 A0A3P9CYY5
Pubmed
EMBL
KK118858
KFM74040.1
MRZV01000519
PIK48493.1
GEZM01079607
GEZM01079598
+ More
JAV62391.1 HADY01023773 HAEJ01006887 SBS47344.1 HADZ01022257 SBP86198.1 ABLF02013930 ABLF02013934 AERX01076149 AERX01070080 GCES01096219 JAQ90103.1 GGMR01000868 MBY13487.1 HAED01014930 SBR01375.1 MRZV01000033 PIK61397.1 GACK01009641 JAA55393.1 MRZV01000161 PIK56894.1 GCES01104253 JAQ82069.1 GEZM01020000 JAV89472.1 GEZM01050997 GEZM01050995 JAV75219.1 GBHO01031825 JAG11779.1 GEZM01051395 JAV74977.1 AGBW02008504 OWR53150.1 MRZV01000718 PIK45343.1 MRZV01000618 PIK46835.1 GFAA01002468 JAU00967.1 ABLF02010707 GGMR01004179 MBY16798.1 GEZM01102055 GEZM01102054 GEZM01102053 GEZM01102051 JAV52011.1 GEGO01004362 JAR91042.1 ABLF02041541 GEZM01097791 JAV54106.1 GEZM01041944 JAV80090.1 MRZV01000576 PIK47528.1 ABLF02038957 GEZM01001973 JAV97431.1 GDKW01000526 JAI56069.1 ABLF02007384 GBHO01036465 JAG07139.1 GEGO01007539 JAR87865.1 ABLF02008456 GBHO01004090 JAG39514.1 MRZV01000723 PIK45255.1 KK122208 KFM82237.1 GFPF01012437 MAA23583.1 GDHC01013957 JAQ04672.1 GEZM01016880 JAV91071.1 KV461054 OCA14957.1 GEZM01066276 JAV68015.1 ABLF02011977 GGMR01014455 MBY27074.1 GEZM01050758 JAV75411.1 ABLF02041865 GBHO01022049 GBHO01022048 JAG21555.1 JAG21556.1 GBBI01004851 JAC13861.1 GEZM01043682 JAV78912.1 HAED01020456 SBR07020.1 GBBI01004852 JAC13860.1 KF319019 AIE48224.1 GEGO01004665 JAR90739.1 ABLF02022355 ABLF02022359 ABLF02022361 ABLF02022365 ABLF02041467 GEZM01001974 JAV97427.1 GFWV01003962 MAA28692.1 ABLF02019772 NCKU01019406 RWR98729.1 GEFM01002416 JAP73380.1 MRZV01001756 PIK36039.1 CM012440 RVE73710.1 LSMT01000199 PFX23692.1 GBHO01013729 JAG29875.1 GBGD01000115 JAC88774.1 ABLF02026678 ABLF02026682
JAV62391.1 HADY01023773 HAEJ01006887 SBS47344.1 HADZ01022257 SBP86198.1 ABLF02013930 ABLF02013934 AERX01076149 AERX01070080 GCES01096219 JAQ90103.1 GGMR01000868 MBY13487.1 HAED01014930 SBR01375.1 MRZV01000033 PIK61397.1 GACK01009641 JAA55393.1 MRZV01000161 PIK56894.1 GCES01104253 JAQ82069.1 GEZM01020000 JAV89472.1 GEZM01050997 GEZM01050995 JAV75219.1 GBHO01031825 JAG11779.1 GEZM01051395 JAV74977.1 AGBW02008504 OWR53150.1 MRZV01000718 PIK45343.1 MRZV01000618 PIK46835.1 GFAA01002468 JAU00967.1 ABLF02010707 GGMR01004179 MBY16798.1 GEZM01102055 GEZM01102054 GEZM01102053 GEZM01102051 JAV52011.1 GEGO01004362 JAR91042.1 ABLF02041541 GEZM01097791 JAV54106.1 GEZM01041944 JAV80090.1 MRZV01000576 PIK47528.1 ABLF02038957 GEZM01001973 JAV97431.1 GDKW01000526 JAI56069.1 ABLF02007384 GBHO01036465 JAG07139.1 GEGO01007539 JAR87865.1 ABLF02008456 GBHO01004090 JAG39514.1 MRZV01000723 PIK45255.1 KK122208 KFM82237.1 GFPF01012437 MAA23583.1 GDHC01013957 JAQ04672.1 GEZM01016880 JAV91071.1 KV461054 OCA14957.1 GEZM01066276 JAV68015.1 ABLF02011977 GGMR01014455 MBY27074.1 GEZM01050758 JAV75411.1 ABLF02041865 GBHO01022049 GBHO01022048 JAG21555.1 JAG21556.1 GBBI01004851 JAC13861.1 GEZM01043682 JAV78912.1 HAED01020456 SBR07020.1 GBBI01004852 JAC13860.1 KF319019 AIE48224.1 GEGO01004665 JAR90739.1 ABLF02022355 ABLF02022359 ABLF02022361 ABLF02022365 ABLF02041467 GEZM01001974 JAV97427.1 GFWV01003962 MAA28692.1 ABLF02019772 NCKU01019406 RWR98729.1 GEFM01002416 JAP73380.1 MRZV01001756 PIK36039.1 CM012440 RVE73710.1 LSMT01000199 PFX23692.1 GBHO01013729 JAG29875.1 GBGD01000115 JAC88774.1 ABLF02026678 ABLF02026682
Proteomes
Pfam
Interpro
IPR021109
Peptidase_aspartic_dom_sf
+ More
IPR000477 RT_dom
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR001878 Znf_CCHC
IPR012337 RNaseH-like_sf
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR001995 Peptidase_A2_cat
IPR005162 Retrotrans_gag_dom
IPR001969 Aspartic_peptidase_AS
IPR038081 CalX-like_sf
IPR036875 Znf_CCHC_sf
IPR041373 RT_RNaseH
IPR000477 RT_dom
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR001878 Znf_CCHC
IPR012337 RNaseH-like_sf
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR001995 Peptidase_A2_cat
IPR005162 Retrotrans_gag_dom
IPR001969 Aspartic_peptidase_AS
IPR038081 CalX-like_sf
IPR036875 Znf_CCHC_sf
IPR041373 RT_RNaseH
Gene 3D
ProteinModelPortal
A0A087U9K1
A0A2G8KKG5
A0A1Y1KPK9
A0A3P8Q6C6
A0A3P9HT22
A0A3B3I1S4
+ More
A0A3B3C9X5 A0A3P8S414 A0A1A8UGA0 A0A1A8D4P4 A0A3P8RHF8 A0A3B1JKR1 A0A3P8RKT7 X1WTY2 A0A3P9M8I3 I3KX76 I3KYE2 A0A146TAZ6 A0A3P9AYU2 A0A2S2N8I3 A0A1A8IVJ0 A0A3P8QAD1 A0A3P8PD24 A0A3B3HLX7 A0A2G8LMB1 A0A3P9HXP1 L7LWW6 A0A2G8L9H5 A0A146SN99 A0A1Y1MUX5 A0A1Y1LNL5 A0A0A9WVK6 A0A1Y1LVD1 A0A212FHD4 A0A3Q0J4U3 A0A2G8KBJ0 A0A2G8KFU3 A0A1E1XPH1 J9LDY4 A0A2S2NI46 A0A1Y1JV53 A0A147BKR8 J9LYD2 A0A1Y1K454 A0A1Y1M671 A0A2G8KHQ8 J9M4C2 A0A1Y1NHV7 A0A0P4VZQ4 J9M6I9 A0A0A9WHB1 A0A147BBY0 J9LNB8 A0A0A9Z4Y4 A0A2G8KB94 A0A087UXZ8 A0A224Z9B6 A0A146LC45 A0A3Q2XZ76 A0A1Y1MZH6 A0A1B8XWD8 A0A1Y1L7V1 J9KCS2 A0A1S3DM59 A0A2S2PCF5 A0A1Y1LR12 X1XQZ7 A0A0A9XW98 A0A023EXW7 A0A1Y1LZ99 A0A1A8JBP0 A0A023EWW4 A0A1S3JEC7 A0A075EIL5 A0A147BIX9 J9L288 A0A1Y1NN34 A0A293LIZ7 J9JQ26 A0A3S3NA82 A0A131Y4S4 A0A2G8JJY8 A0A2I4C5Y1 A0A3S2N4F0 A0A2B4S589 A0A0A9YK20 A0A1S3ICH8 A0A069DZS8 J9L8Q1 A0A3P9CYY5
A0A3B3C9X5 A0A3P8S414 A0A1A8UGA0 A0A1A8D4P4 A0A3P8RHF8 A0A3B1JKR1 A0A3P8RKT7 X1WTY2 A0A3P9M8I3 I3KX76 I3KYE2 A0A146TAZ6 A0A3P9AYU2 A0A2S2N8I3 A0A1A8IVJ0 A0A3P8QAD1 A0A3P8PD24 A0A3B3HLX7 A0A2G8LMB1 A0A3P9HXP1 L7LWW6 A0A2G8L9H5 A0A146SN99 A0A1Y1MUX5 A0A1Y1LNL5 A0A0A9WVK6 A0A1Y1LVD1 A0A212FHD4 A0A3Q0J4U3 A0A2G8KBJ0 A0A2G8KFU3 A0A1E1XPH1 J9LDY4 A0A2S2NI46 A0A1Y1JV53 A0A147BKR8 J9LYD2 A0A1Y1K454 A0A1Y1M671 A0A2G8KHQ8 J9M4C2 A0A1Y1NHV7 A0A0P4VZQ4 J9M6I9 A0A0A9WHB1 A0A147BBY0 J9LNB8 A0A0A9Z4Y4 A0A2G8KB94 A0A087UXZ8 A0A224Z9B6 A0A146LC45 A0A3Q2XZ76 A0A1Y1MZH6 A0A1B8XWD8 A0A1Y1L7V1 J9KCS2 A0A1S3DM59 A0A2S2PCF5 A0A1Y1LR12 X1XQZ7 A0A0A9XW98 A0A023EXW7 A0A1Y1LZ99 A0A1A8JBP0 A0A023EWW4 A0A1S3JEC7 A0A075EIL5 A0A147BIX9 J9L288 A0A1Y1NN34 A0A293LIZ7 J9JQ26 A0A3S3NA82 A0A131Y4S4 A0A2G8JJY8 A0A2I4C5Y1 A0A3S2N4F0 A0A2B4S589 A0A0A9YK20 A0A1S3ICH8 A0A069DZS8 J9L8Q1 A0A3P9CYY5
Ontologies
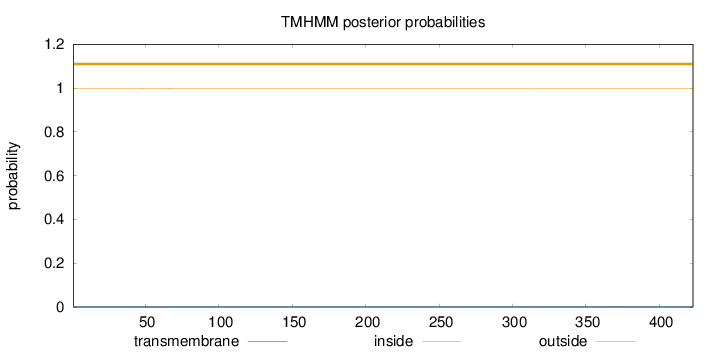
Topology
Length:
423
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02207
Exp number, first 60 AAs:
0.0129
Total prob of N-in:
0.00111
outside
1 - 423
Population Genetic Test Statistics
Pi
3.375172
Theta
8.309396
Tajima's D
-1.753761
CLR
39.690405
CSRT
0.0283485825708715
Interpretation
Uncertain
