Gene
KWMTBOMO14203
Pre Gene Modal
BGIBMGA011140
Annotation
PREDICTED:_fatty_acyl-CoA_reductase_1-like_[Bombyx_mori]
Full name
Fatty acyl-CoA reductase
Location in the cell
Cytoplasmic Reliability : 1.432 Mitochondrial Reliability : 1.705
Sequence
CDS
ATGGACAGCGCTTTGGCTTTGGAGATCGAGCAGGTCTCGCGTCAGAAGGCGCTGGAGGAGGAAGCGCTTCGCGGCGAGTCGGCTGTGCGGCAGTTCTACGCGGGAACCACAGCGCTGATCACTGGAGGGACGGGCTTCATCGGGAAACAACTCATAGAGAAGCTTCTAAGGTCGTGCGACGTTAAACGAATTTACCTGTTGATTAGGTCGAAGAAAGGGAAATCGATGCAAGAGAGACTAAAGTCACAATTAAATGATCCAGTATACGACAATTTACGGTCTCAACAACCAGATTTCACTCGGAAAGTTATACCGGTGGAAGGCGACATCTCAGAACTAGGCGTCGGCCTGAACAACGAAGACAGGATGAAGCTTGCGCAAGAGGTGAATGTGATATTCCATGTGGCTGCCACGACCAGGTTCGAAGAGCCACTGAAGAAGGCCACATTCATCAACGTCAGAGGAACCAGGGAGATCATGGAATTGGCCAAATGCTGCAAAGATCTAAGGTCCTTCGTACACGTGTCAACTGCGTTCTCTAATGCAACCAAGAGCAGAATAAATCACGATTTGGAAGAGAAAATCTATGAAAGCCCTGTCCCTCCGGAAGTCATGATACATCTCGCGGAGACAGTCGAAGAATGCAGACTCAACGCGATCACTGCTGATTTGATAAGGGACTGGCCCAACACGTACTGTTTCACGAAGGCCATAGCCGAGGATGTCGTGCGTACAATGGGCAAGGATTTACCTGTGGCCATAGTGAGGCCACCCGTAGTTATCTGCACGCTCAATGAACCCACTCCTGGATGGGTGGATATGAGCTCGGTGATCGGTCCTAGTGGGGTAAGTGCAAATCAAAGTTGGGACTTGTTAATAGCGGTTCGCACAGTGTACATAGTTGGGGTTTATTTTAAGATTTTTAAGCACACCCATGGTAACACCTTACTGCCCAATTCTCAAGCGAAGTAA
Protein
MDSALALEIEQVSRQKALEEEALRGESAVRQFYAGTTALITGGTGFIGKQLIEKLLRSCDVKRIYLLIRSKKGKSMQERLKSQLNDPVYDNLRSQQPDFTRKVIPVEGDISELGVGLNNEDRMKLAQEVNVIFHVAATTRFEEPLKKATFINVRGTREIMELAKCCKDLRSFVHVSTAFSNATKSRINHDLEEKIYESPVPPEVMIHLAETVEECRLNAITADLIRDWPNTYCFTKAIAEDVVRTMGKDLPVAIVRPPVVICTLNEPTPGWVDMSSVIGPSGVSANQSWDLLIAVRTVYIVGVYFKIFKHTHGNTLLPNSQAK
Summary
Description
Catalyzes the reduction of fatty acyl-CoA to fatty alcohols.
Catalytic Activity
a long-chain fatty acyl-CoA + 2 H(+) + 2 NADPH = a long-chain primary fatty alcohol + CoA + 2 NADP(+)
Similarity
Belongs to the fatty acyl-CoA reductase family.
Uniprot
H9JNN6
H9JNM2
H9JWY2
H9JNN4
A0A2A4K1U6
A0A2A4JYT4
+ More
A0A2A4K1F7 A0A2A4JY42 A0A2H1VWV0 A0A2W1B8Q0 A0A2A4K2D3 A0A2W1BH40 A0A2H1VAJ7 A0A2A4JY41 A0A2A4J5Y0 A0A2W1BUR6 A0A2W1BRZ8 A0A0P0ELA9 A0A2A4JIF6 A0A2W1BEZ6 A0A291P0Q6 A0A212ER01 A0A2W1BFX3 A0A2H1WT26 A0A194R4R4 A0A194PZ57 A0A194PSN1 A0A0N1PK47 A0A2A4JG81 A0A194PT03 A0A2H1VVN7 U5KFQ5 A0A194PV95 A0A0P0EM43 A0A1V0M874 A0A0F6Q1J7 A0A2W1BH51 A0A0F6Q2N3 A0A194PTV2 A0A194QZH1 A0A068FK94 A0A2A4JS90 A0A2H1VVB2 H9JNP1 A0A0N1I9W9 A0A2J7RN95 A0A212F0D5 A0A067R2Y2 U4UBB5 N6TBL5 A0A2J7RNB9 A0A195BRC5 A0A158P2S4 K7J6N4 A0A3L8DXX4 A0A232EYC4 A0A195DFI7 A0A151X2I2 A0A193CHN6 A0A1J1J054 K7J6N1 F4W5T8 A0A195C2N5 A0A232F2D0 A0A1Y1LHX9 A0A336L0D6 A0A1B0GID8 A0A195EZP6 E2AWD8 A0A1B6DYP0 E9I8G2 A0A310S4M5 E2BCH8 A0A069DU52 E2AKQ3 A0A0L0BW16 A0A0M9ABB1 A0A3Q8FLY7 A0A0A9WBX3 A0A2D0T2W8 A0A0C9PMV0 A0A1B0CRM9 A0A2D0T371 T1HHV9 A0A1I8P794 A0A2J7QR94 A0A1B6EEG7 W5U6W2 A0A0L7RAS9 A0A0A9XGI9 A0A158NHD8 D2A5T7 A0A146LDG8 E2AWD9 K7J7I4
A0A2A4K1F7 A0A2A4JY42 A0A2H1VWV0 A0A2W1B8Q0 A0A2A4K2D3 A0A2W1BH40 A0A2H1VAJ7 A0A2A4JY41 A0A2A4J5Y0 A0A2W1BUR6 A0A2W1BRZ8 A0A0P0ELA9 A0A2A4JIF6 A0A2W1BEZ6 A0A291P0Q6 A0A212ER01 A0A2W1BFX3 A0A2H1WT26 A0A194R4R4 A0A194PZ57 A0A194PSN1 A0A0N1PK47 A0A2A4JG81 A0A194PT03 A0A2H1VVN7 U5KFQ5 A0A194PV95 A0A0P0EM43 A0A1V0M874 A0A0F6Q1J7 A0A2W1BH51 A0A0F6Q2N3 A0A194PTV2 A0A194QZH1 A0A068FK94 A0A2A4JS90 A0A2H1VVB2 H9JNP1 A0A0N1I9W9 A0A2J7RN95 A0A212F0D5 A0A067R2Y2 U4UBB5 N6TBL5 A0A2J7RNB9 A0A195BRC5 A0A158P2S4 K7J6N4 A0A3L8DXX4 A0A232EYC4 A0A195DFI7 A0A151X2I2 A0A193CHN6 A0A1J1J054 K7J6N1 F4W5T8 A0A195C2N5 A0A232F2D0 A0A1Y1LHX9 A0A336L0D6 A0A1B0GID8 A0A195EZP6 E2AWD8 A0A1B6DYP0 E9I8G2 A0A310S4M5 E2BCH8 A0A069DU52 E2AKQ3 A0A0L0BW16 A0A0M9ABB1 A0A3Q8FLY7 A0A0A9WBX3 A0A2D0T2W8 A0A0C9PMV0 A0A1B0CRM9 A0A2D0T371 T1HHV9 A0A1I8P794 A0A2J7QR94 A0A1B6EEG7 W5U6W2 A0A0L7RAS9 A0A0A9XGI9 A0A158NHD8 D2A5T7 A0A146LDG8 E2AWD9 K7J7I4
EC Number
1.2.1.84
Pubmed
EMBL
BABH01035122
BABH01035123
BABH01035124
BABH01035129
BABH01035130
BABH01035131
+ More
BABH01035132 BABH01042202 BABH01042203 BABH01042204 BABH01035110 NWSH01000263 PCG77898.1 NWSH01000368 PCG76936.1 PCG77899.1 PCG76935.1 ODYU01004617 SOQ44684.1 KZ150217 PZC72129.1 NWSH01000230 PCG78176.1 PZC72126.1 ODYU01001262 SOQ37234.1 PCG76937.1 NWSH01003107 PCG66934.1 KZ149959 PZC76396.1 PZC76395.1 KT261699 ALJ30239.1 NWSH01001446 PCG71213.1 KZ150147 PZC72871.1 MF687542 ATJ44468.1 AGBW02013183 OWR43905.1 PZC72127.1 ODYU01010853 SOQ56210.1 KQ460890 KPJ10851.1 KQ459593 KPI96440.1 KPI96441.1 KQ460036 KPJ18429.1 NWSH01001490 PCG71095.1 KPI96442.1 ODYU01004416 SOQ44264.1 JX989158 AGR49317.1 KPI96679.1 KT261695 ALJ30235.1 KU755476 ARD71186.1 KP237926 MF687596 AKD01779.1 ATJ44522.1 PZC72130.1 KP237913 MF687534 AKD01766.1 ATJ44460.1 KPI96403.1 KPJ10852.1 KJ622061 AID66649.1 NWSH01000716 PCG74636.1 ODYU01004649 SOQ44753.1 BABH01035163 BABH01035164 KPJ18430.1 NEVH01002545 PNF42310.1 AGBW02011118 OWR47192.1 KK852740 KDR17399.1 KB632003 ERL87876.1 APGK01044237 APGK01044238 KB741021 ENN75098.1 PNF42309.1 KQ976417 KYM89582.1 ADTU01007436 ADTU01007437 QOIP01000002 RLU25314.1 NNAY01001641 OXU23345.1 KQ980903 KYN11612.1 KQ982576 KYQ54647.1 KU963289 ANN23959.1 CVRI01000065 CRL05837.1 GL887695 EGI70414.1 KQ978379 KYM94471.1 NNAY01001243 OXU24628.1 GEZM01061226 JAV70627.1 UFQS01001285 UFQT01001285 SSX10116.1 SSX29838.1 AJWK01013757 KQ981905 KYN33349.1 GL443286 EFN62238.1 GEDC01006523 JAS30775.1 GL761538 EFZ23139.1 KQ776822 OAD52165.1 GL447283 EFN86604.1 GBGD01001246 JAC87643.1 GL440368 EFN65993.1 JRES01001250 KNC24227.1 KQ435694 KOX80980.1 MG573160 AWJ25022.1 GBHO01038325 GBHO01038321 GBRD01012464 GDHC01011251 JAG05279.1 JAG05283.1 JAG53360.1 JAQ07378.1 GBYB01002483 JAG72250.1 AJWK01024997 AJWK01024998 AJWK01024999 ACPB03001017 NEVH01011924 PNF31087.1 GEDC01000976 JAS36322.1 JT406763 AHH37716.1 KQ414617 KOC68027.1 GBHO01025676 JAG17928.1 ADTU01015701 ADTU01015702 KQ971345 EFA05024.1 GDHC01012276 JAQ06353.1 EFN62239.1
BABH01035132 BABH01042202 BABH01042203 BABH01042204 BABH01035110 NWSH01000263 PCG77898.1 NWSH01000368 PCG76936.1 PCG77899.1 PCG76935.1 ODYU01004617 SOQ44684.1 KZ150217 PZC72129.1 NWSH01000230 PCG78176.1 PZC72126.1 ODYU01001262 SOQ37234.1 PCG76937.1 NWSH01003107 PCG66934.1 KZ149959 PZC76396.1 PZC76395.1 KT261699 ALJ30239.1 NWSH01001446 PCG71213.1 KZ150147 PZC72871.1 MF687542 ATJ44468.1 AGBW02013183 OWR43905.1 PZC72127.1 ODYU01010853 SOQ56210.1 KQ460890 KPJ10851.1 KQ459593 KPI96440.1 KPI96441.1 KQ460036 KPJ18429.1 NWSH01001490 PCG71095.1 KPI96442.1 ODYU01004416 SOQ44264.1 JX989158 AGR49317.1 KPI96679.1 KT261695 ALJ30235.1 KU755476 ARD71186.1 KP237926 MF687596 AKD01779.1 ATJ44522.1 PZC72130.1 KP237913 MF687534 AKD01766.1 ATJ44460.1 KPI96403.1 KPJ10852.1 KJ622061 AID66649.1 NWSH01000716 PCG74636.1 ODYU01004649 SOQ44753.1 BABH01035163 BABH01035164 KPJ18430.1 NEVH01002545 PNF42310.1 AGBW02011118 OWR47192.1 KK852740 KDR17399.1 KB632003 ERL87876.1 APGK01044237 APGK01044238 KB741021 ENN75098.1 PNF42309.1 KQ976417 KYM89582.1 ADTU01007436 ADTU01007437 QOIP01000002 RLU25314.1 NNAY01001641 OXU23345.1 KQ980903 KYN11612.1 KQ982576 KYQ54647.1 KU963289 ANN23959.1 CVRI01000065 CRL05837.1 GL887695 EGI70414.1 KQ978379 KYM94471.1 NNAY01001243 OXU24628.1 GEZM01061226 JAV70627.1 UFQS01001285 UFQT01001285 SSX10116.1 SSX29838.1 AJWK01013757 KQ981905 KYN33349.1 GL443286 EFN62238.1 GEDC01006523 JAS30775.1 GL761538 EFZ23139.1 KQ776822 OAD52165.1 GL447283 EFN86604.1 GBGD01001246 JAC87643.1 GL440368 EFN65993.1 JRES01001250 KNC24227.1 KQ435694 KOX80980.1 MG573160 AWJ25022.1 GBHO01038325 GBHO01038321 GBRD01012464 GDHC01011251 JAG05279.1 JAG05283.1 JAG53360.1 JAQ07378.1 GBYB01002483 JAG72250.1 AJWK01024997 AJWK01024998 AJWK01024999 ACPB03001017 NEVH01011924 PNF31087.1 GEDC01000976 JAS36322.1 JT406763 AHH37716.1 KQ414617 KOC68027.1 GBHO01025676 JAG17928.1 ADTU01015701 ADTU01015702 KQ971345 EFA05024.1 GDHC01012276 JAQ06353.1 EFN62239.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000235965
+ More
UP000027135 UP000030742 UP000019118 UP000078540 UP000005205 UP000002358 UP000279307 UP000215335 UP000078492 UP000075809 UP000183832 UP000007755 UP000078542 UP000092461 UP000078541 UP000000311 UP000008237 UP000037069 UP000053105 UP000221080 UP000015103 UP000095300 UP000053825 UP000007266
UP000027135 UP000030742 UP000019118 UP000078540 UP000005205 UP000002358 UP000279307 UP000215335 UP000078492 UP000075809 UP000183832 UP000007755 UP000078542 UP000092461 UP000078541 UP000000311 UP000008237 UP000037069 UP000053105 UP000221080 UP000015103 UP000095300 UP000053825 UP000007266
Interpro
SUPFAM
SSF51735
SSF51735
CDD
ProteinModelPortal
H9JNN6
H9JNM2
H9JWY2
H9JNN4
A0A2A4K1U6
A0A2A4JYT4
+ More
A0A2A4K1F7 A0A2A4JY42 A0A2H1VWV0 A0A2W1B8Q0 A0A2A4K2D3 A0A2W1BH40 A0A2H1VAJ7 A0A2A4JY41 A0A2A4J5Y0 A0A2W1BUR6 A0A2W1BRZ8 A0A0P0ELA9 A0A2A4JIF6 A0A2W1BEZ6 A0A291P0Q6 A0A212ER01 A0A2W1BFX3 A0A2H1WT26 A0A194R4R4 A0A194PZ57 A0A194PSN1 A0A0N1PK47 A0A2A4JG81 A0A194PT03 A0A2H1VVN7 U5KFQ5 A0A194PV95 A0A0P0EM43 A0A1V0M874 A0A0F6Q1J7 A0A2W1BH51 A0A0F6Q2N3 A0A194PTV2 A0A194QZH1 A0A068FK94 A0A2A4JS90 A0A2H1VVB2 H9JNP1 A0A0N1I9W9 A0A2J7RN95 A0A212F0D5 A0A067R2Y2 U4UBB5 N6TBL5 A0A2J7RNB9 A0A195BRC5 A0A158P2S4 K7J6N4 A0A3L8DXX4 A0A232EYC4 A0A195DFI7 A0A151X2I2 A0A193CHN6 A0A1J1J054 K7J6N1 F4W5T8 A0A195C2N5 A0A232F2D0 A0A1Y1LHX9 A0A336L0D6 A0A1B0GID8 A0A195EZP6 E2AWD8 A0A1B6DYP0 E9I8G2 A0A310S4M5 E2BCH8 A0A069DU52 E2AKQ3 A0A0L0BW16 A0A0M9ABB1 A0A3Q8FLY7 A0A0A9WBX3 A0A2D0T2W8 A0A0C9PMV0 A0A1B0CRM9 A0A2D0T371 T1HHV9 A0A1I8P794 A0A2J7QR94 A0A1B6EEG7 W5U6W2 A0A0L7RAS9 A0A0A9XGI9 A0A158NHD8 D2A5T7 A0A146LDG8 E2AWD9 K7J7I4
A0A2A4K1F7 A0A2A4JY42 A0A2H1VWV0 A0A2W1B8Q0 A0A2A4K2D3 A0A2W1BH40 A0A2H1VAJ7 A0A2A4JY41 A0A2A4J5Y0 A0A2W1BUR6 A0A2W1BRZ8 A0A0P0ELA9 A0A2A4JIF6 A0A2W1BEZ6 A0A291P0Q6 A0A212ER01 A0A2W1BFX3 A0A2H1WT26 A0A194R4R4 A0A194PZ57 A0A194PSN1 A0A0N1PK47 A0A2A4JG81 A0A194PT03 A0A2H1VVN7 U5KFQ5 A0A194PV95 A0A0P0EM43 A0A1V0M874 A0A0F6Q1J7 A0A2W1BH51 A0A0F6Q2N3 A0A194PTV2 A0A194QZH1 A0A068FK94 A0A2A4JS90 A0A2H1VVB2 H9JNP1 A0A0N1I9W9 A0A2J7RN95 A0A212F0D5 A0A067R2Y2 U4UBB5 N6TBL5 A0A2J7RNB9 A0A195BRC5 A0A158P2S4 K7J6N4 A0A3L8DXX4 A0A232EYC4 A0A195DFI7 A0A151X2I2 A0A193CHN6 A0A1J1J054 K7J6N1 F4W5T8 A0A195C2N5 A0A232F2D0 A0A1Y1LHX9 A0A336L0D6 A0A1B0GID8 A0A195EZP6 E2AWD8 A0A1B6DYP0 E9I8G2 A0A310S4M5 E2BCH8 A0A069DU52 E2AKQ3 A0A0L0BW16 A0A0M9ABB1 A0A3Q8FLY7 A0A0A9WBX3 A0A2D0T2W8 A0A0C9PMV0 A0A1B0CRM9 A0A2D0T371 T1HHV9 A0A1I8P794 A0A2J7QR94 A0A1B6EEG7 W5U6W2 A0A0L7RAS9 A0A0A9XGI9 A0A158NHD8 D2A5T7 A0A146LDG8 E2AWD9 K7J7I4
PDB
4W4T
E-value=8.37677e-11,
Score=160
Ontologies
PANTHER
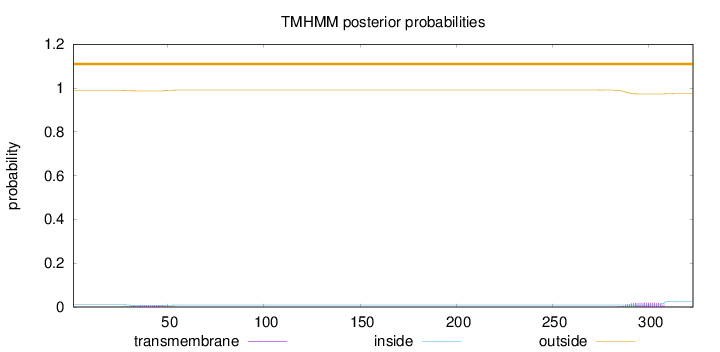
Topology
Length:
323
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.51029
Exp number, first 60 AAs:
0.14347
Total prob of N-in:
0.01033
outside
1 - 323
Population Genetic Test Statistics
Pi
19.340922
Theta
22.526961
Tajima's D
-0.503088
CLR
0.272552
CSRT
0.24058797060147
Interpretation
Uncertain
