Gene
KWMTBOMO14190
Annotation
reverse_transcriptase_[Bombyx_mori]
Full name
RNA-directed DNA polymerase from mobile element jockey
+ More
Probable RNA-directed DNA polymerase from transposon BS
Probable RNA-directed DNA polymerase from transposon BS
Alternative Name
Reverse transcriptase
Location in the cell
Nuclear Reliability : 2.41
Sequence
CDS
ATGGCGCAAAACGCTAGAGAGAAACCATATTCCCTTACGTTAGCATTCTACAATGCTAACGGACTCGCGCGACAACGCGATCAAATTTACGAATTCCTCCGCGACAATCTTGTAGATATTCTGCTAGTGCAGGAGACCTGCCTGAAGCCCTCGCGTCGTGACCCGAAAGTCGCGAATTACGTCATGGTTAGGAATGACAGACTCACCGCCTCCAAAGGCGGGACTGCCATTTACTATAGGCGGGCCCTGCACGTTGTCCCTCTCGATACCCCCTCGCTCTCACATATCGAGGCGTCAGTGTGCCGTATCTCGCTGACGGGACACCAGCCGATCGTCATCGCATCCGTCTATCTCCCCCCGGACAAGCCCCTTCTGAGCAGTGACATCGAGTCACTGTTCGGCATGGGAGACTCCGTCATCCTGGCAGGCGATTTAAATTGCCACCACACTAGGTGGAACTGCCACAGTTCAAACATAAATGGTAGGCGTCTCGACGCGTTTATAGACGACCTTACCTTTGAAGTAGTCGGTCCCCCAACTCCAACATGTTATCCGTATAACATCGCGCTCCGTCCGAGCACTATAGACCTGGCACTGCTGAGGAACGTAACTCTGCGCTTGCGTTCCATCGAAGCAATTCACAGCTACGAGGACCATGGTGGACTGGAAGAAGCTGGGCAAATGCTTAGCCGATGCCGCTCCACCAATCCTCCCTGCGGCCCGGATTCAACTCCCTCCCCCGAGGACACAGTCGAATCCATAAACATCATCACGGATCACATCTCTTCCGCGATCATAAGATCCTCAAAAGAAGTCGATGTGGAGGATAGCTTCCACCGCATCAGACTGTCCCCCGAACTTAGGAACCTCCTAAGAGTTAGAAACGCGGCAATCCGGGCCTACGATCAACTTCCCACGCACTCAAACCGGATTCGGATGCGTCGTCTACAGCGGGATGTAAAATCTCGCCTAAGCGACGCCCGAAATGAGAATTGGGATAGTTACTTAGAAGGACTCGCGCCCTCTCATCAAGCATACTGGCGACTAGCAAGGACTCTCAAGTCCGAAACTACCGCTACTATGCCTCCTCTCGTACGCCCTTCAGGCCAACCACCGGCTTTCGATGACGATGACAAGGCAGAGTTGCTGGCCGATGCACTGCAAGAGCAGTGCACCACCAGCACTCAATACGCGGACCCCGAACACACCGAGGCAGTCGACAGGGAGGTCGAGCGCAGAGCTTCCCTGCCACCCTCGGACGCGTTACCCCCCATTACCTCTGACGAAGTTAGAGAAGTAATAGACAACCTTCAACCTAAGAAGGCACCCGGCTACGACGGCATCCGCAACCGCGCGTTAAAACTCTTACCAGTCCAACTGATAACAATGTTGGCTACCATCTTAAATGCTGCTATGACGCACTGCATCTTTCCGGCGGTATGGAAAGAAGCAGACGTTATCGGTATACACAAGCCGGGCAAACCGAATAATGAAACCGCTAGTTACCGACCGATTAGTCTCCTCCCAGCGATAGGCAAAATTTACGAACGGCTCCTTCGGAAACGCCTCTGGGACTTCGTTACCGCGAATAAAATTCTAATAGACGAGCAGTTTGGATTCCGCGCCAAACACTCGTGCGTACAACAAGTGCACCGCCTCACGGAGCACATCTTAATAGGGCTAAATAGGCGTAAACAAATCCCGACCGGCGCCCTCTTCTTCGATGTAGCGAAGGCGTTCGACAAAGTCTGGCACAACGGTTTGATCTACAAACTGTACAACATGGGAGTGCCAGACAGACTCGTGCTCATCATACGAGACTTCTTGTCGAACCGTTCGTTTCGATATCGAGTAGAGGGAACTCGTTCTCGTCCCCGTCATCTGACTGCCGGAGTCCCGCAAGGCTCCGCACTCTCCCCGTTACTATTTAGTTTGTATATCAACGATATACCCCGGTCTCCGGAGACCCATCTAGCGCTCTTCGCCGATGACACGGCTATCTACTACTCGTGTAGGAAGATGTCGTTGCTTCATCGGCGACTCCAGACCGCAGTAACCACCATGGGACAGTGGTTCCGGAAGTGGCGAATAGACATTAACCCCACGAAAAGCACAGCGGTGCTATTCAAAAGGGGTCGCCCTCCGAATACCACTTCGAGCACCCCACTCCCCAGTAGGCGCGTAAACACCTCCGCCGTTAGCCCCATCACTCTCTTTGACCAGCCCATACCGTGGGCCCCGAAGGTCAAATACCTAGGCGTCACCCTCGACAGAGGGATGACATTCCGTCCCCACGTAAAAACGGTACGCGATCGCGCCGCGTTTATACTAGGACGACTCTACCCAATGCTTTGTAGACGAAGCAAACTGTCCCTCCGTAACAAGGTAACCCTCTACAAAACTTGCATACGCCCCGTCATGACGTATGCAAGCGTAGTGTTCGCTCACGCGGCCCGCACCAACTTGAAACCCCTTCAGGTAATACAATCCCGATTCTGCAGGATAGCCGTCGGAGCACCATGGTTCCAGAGGAACGTGGACCTCCACGATGACCTGGAGCTCGACCCCGTCAGTAAGTATCTACAGTCGGCATCGCTGCGCCACTTTGAGAAGGCGGCACGACATGAGAACCCTCTTGTCGTGGCCGCCGGAAACTACATACCCGATCCTGTAGACCGTTTGCTAAACATCCGAAAAAGGCTCTCTTCCATCTGCCGGGAACGTGTTATGAGCTTCTTCGGACACATCATGCGGTCCGCCAGATATGAATTAGAGAAGCTCATAATTGCGGGTCGCATAGAGGGCAAGCGCGCCGTGGGCAGAACACCAGCCAGATAG
Protein
MAQNAREKPYSLTLAFYNANGLARQRDQIYEFLRDNLVDILLVQETCLKPSRRDPKVANYVMVRNDRLTASKGGTAIYYRRALHVVPLDTPSLSHIEASVCRISLTGHQPIVIASVYLPPDKPLLSSDIESLFGMGDSVILAGDLNCHHTRWNCHSSNINGRRLDAFIDDLTFEVVGPPTPTCYPYNIALRPSTIDLALLRNVTLRLRSIEAIHSYEDHGGLEEAGQMLSRCRSTNPPCGPDSTPSPEDTVESINIITDHISSAIIRSSKEVDVEDSFHRIRLSPELRNLLRVRNAAIRAYDQLPTHSNRIRMRRLQRDVKSRLSDARNENWDSYLEGLAPSHQAYWRLARTLKSETTATMPPLVRPSGQPPAFDDDDKAELLADALQEQCTTSTQYADPEHTEAVDREVERRASLPPSDALPPITSDEVREVIDNLQPKKAPGYDGIRNRALKLLPVQLITMLATILNAAMTHCIFPAVWKEADVIGIHKPGKPNNETASYRPISLLPAIGKIYERLLRKRLWDFVTANKILIDEQFGFRAKHSCVQQVHRLTEHILIGLNRRKQIPTGALFFDVAKAFDKVWHNGLIYKLYNMGVPDRLVLIIRDFLSNRSFRYRVEGTRSRPRHLTAGVPQGSALSPLLFSLYINDIPRSPETHLALFADDTAIYYSCRKMSLLHRRLQTAVTTMGQWFRKWRIDINPTKSTAVLFKRGRPPNTTSSTPLPSRRVNTSAVSPITLFDQPIPWAPKVKYLGVTLDRGMTFRPHVKTVRDRAAFILGRLYPMLCRRSKLSLRNKVTLYKTCIRPVMTYASVVFAHAARTNLKPLQVIQSRFCRIAVGAPWFQRNVDLHDDLELDPVSKYLQSASLRHFEKAARHENPLVVAAGNYIPDPVDRLLNIRKRLSSICRERVMSFFGHIMRSARYELEKLIIAGRIEGKRAVGRTPAR
Summary
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Cofactor
Mg(2+)
Mn(2+)
Mn(2+)
Miscellaneous
Prefers poly(rC) and poly(rA) as template and activated DNA is not effective.
Keywords
Nucleotidyltransferase
RNA-directed DNA polymerase
Transferase
Transposable element
Feature
chain RNA-directed DNA polymerase from mobile element jockey
Uniprot
Q93137
Q9BPQ0
Q9XXW0
A0A0N1PFV1
O96546
A0A1Y1NA23
+ More
A0A139W8G9 A0A139WA11 V5GNF3 V5GNJ2 A0A023EYS8 A0A3N5Z7I3 A0A023EZE6 D7EM17 Q07995 A0A2S2PPK8 X1WLB2 A0A2S2Q383 A0A2M4CV42 A0A2M4CV52 A0A2S2QEE0 X1WNI2 A0A2M4BD66 A0A2M4BDA5 A0A2M4BD62 A0A087UGF5 A0A224XF24 A0A2J7R7D0 A0A2M4BD67 A0A2S2NGV8 A0A2M4BDV6 A0A2J7PU96 A0A224X6S1 A0A224X6Q4 A0A2S2N814 A0A2J7QR37 A0A224X6T4 A0A224XI99 A0A2J7PBY3 A0A224X6U5 A0A2J7QY69 A0A2S2P698 A0A224XB44 A0A069DZL8 A0A2J7QB68 X1WQI9 A0A2H8TF37 J9LC21 X1WZH0 A0A224X6R6 J9LFC9 A0A2J7Q6J2 J9M522 A0A2S2R9D4 A0A2M4BD83 A0A2S2NPG3 A0A2J7PVX3 A0A2H8TKI9 A0A2M4CVF1 A0A2S2PHL7 A0A2M4CVK2 A0A224X746 A0A0A9YJ00 A0A224X6W2 A0A2S2P6C4 A0A0A9YU04 A0A2M4BEF4 A0A2S2PJG2 A0A2M4BEZ9 A0A1Y1LKN6 A0A2J7PDM0 P21328 A0A2A4IUM2 A0A224XHW8 K7IN67 A0A069DX78 A0A2S2NH02 Q95SX7 Q04135 A0A224XJI0 A0A2S2PA10 A0A1Y1KFR3 A0A142LX26 A0A352X8G4 Q6R811 P21329 A0A2J7RD70 Q961V7 A0A2S2N6Q7 J9KLB1 A0A0A9YSX4 A0A2S2P6S3
A0A139W8G9 A0A139WA11 V5GNF3 V5GNJ2 A0A023EYS8 A0A3N5Z7I3 A0A023EZE6 D7EM17 Q07995 A0A2S2PPK8 X1WLB2 A0A2S2Q383 A0A2M4CV42 A0A2M4CV52 A0A2S2QEE0 X1WNI2 A0A2M4BD66 A0A2M4BDA5 A0A2M4BD62 A0A087UGF5 A0A224XF24 A0A2J7R7D0 A0A2M4BD67 A0A2S2NGV8 A0A2M4BDV6 A0A2J7PU96 A0A224X6S1 A0A224X6Q4 A0A2S2N814 A0A2J7QR37 A0A224X6T4 A0A224XI99 A0A2J7PBY3 A0A224X6U5 A0A2J7QY69 A0A2S2P698 A0A224XB44 A0A069DZL8 A0A2J7QB68 X1WQI9 A0A2H8TF37 J9LC21 X1WZH0 A0A224X6R6 J9LFC9 A0A2J7Q6J2 J9M522 A0A2S2R9D4 A0A2M4BD83 A0A2S2NPG3 A0A2J7PVX3 A0A2H8TKI9 A0A2M4CVF1 A0A2S2PHL7 A0A2M4CVK2 A0A224X746 A0A0A9YJ00 A0A224X6W2 A0A2S2P6C4 A0A0A9YU04 A0A2M4BEF4 A0A2S2PJG2 A0A2M4BEZ9 A0A1Y1LKN6 A0A2J7PDM0 P21328 A0A2A4IUM2 A0A224XHW8 K7IN67 A0A069DX78 A0A2S2NH02 Q95SX7 Q04135 A0A224XJI0 A0A2S2PA10 A0A1Y1KFR3 A0A142LX26 A0A352X8G4 Q6R811 P21329 A0A2J7RD70 Q961V7 A0A2S2N6Q7 J9KLB1 A0A0A9YSX4 A0A2S2P6S3
EC Number
2.7.7.49
Pubmed
EMBL
U07847
AAA17752.1
AB055391
BAB21761.1
AB018558
BAA76304.1
+ More
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 KQ973342 KXZ75569.1 KQ971729 KYB24751.1 GALX01005359 JAB63107.1 GALX01005299 JAB63167.1 GBBI01004395 JAC14317.1 RPOV01000134 RPJ78669.1 GBBI01004571 JAC14141.1 KQ973412 EFA12510.2 S59870 AAB26437.2 GGMR01018716 MBY31335.1 ABLF02042518 ABLF02057746 GGMS01002992 MBY72195.1 GGFL01005012 MBW69190.1 GGFL01004957 MBW69135.1 GGMS01006359 MBY75562.1 ABLF02008153 ABLF02008462 ABLF02008464 GGFJ01001844 MBW50985.1 GGFJ01001842 MBW50983.1 GGFJ01001843 MBW50984.1 KK119692 KFM76444.1 GFTR01008038 JAW08388.1 NEVH01006736 PNF36716.1 GGFJ01001858 MBW50999.1 GGMR01003407 MBY16026.1 GGFJ01001857 MBW50998.1 NEVH01021205 PNF19912.1 GFTR01008271 JAW08155.1 GFTR01008279 JAW08147.1 GGMR01000676 MBY13295.1 NEVH01011985 PNF31054.1 GFTR01008261 JAW08165.1 GFTR01008256 JAW08170.1 NEVH01027074 PNF13841.1 GFTR01008251 JAW08175.1 NEVH01009134 PNF33528.1 GGMR01012305 MBY24924.1 GFTR01008262 JAW08164.1 GBGD01000415 JAC88474.1 NEVH01016302 PNF25823.1 ABLF02022418 GFXV01000908 MBW12713.1 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 ABLF02034467 GFTR01008269 JAW08157.1 ABLF02023911 NEVH01017470 PNF24199.1 ABLF02024246 GGMS01017365 MBY86568.1 GGFJ01001822 MBW50963.1 GGMR01006408 MBY19027.1 NEVH01020940 PNF20481.1 GFXV01002840 MBW14645.1 GGFL01005122 MBW69300.1 GGMR01016296 MBY28915.1 GGFL01005121 MBW69299.1 GFTR01008151 JAW08275.1 GBHO01010547 GBHO01010546 JAG33057.1 JAG33058.1 GFTR01008260 JAW08166.1 GGMR01012283 MBY24902.1 GBHO01021814 GBHO01010549 JAG21790.1 JAG33055.1 GGFJ01002256 MBW51397.1 GGMR01016905 MBY29524.1 GGFJ01002257 MBW51398.1 GEZM01053022 JAV74219.1 NEVH01026386 PNF14434.1 M22874 M38643 NWSH01007753 PCG62823.1 GFTR01008254 JAW08172.1 GBGD01000389 JAC88500.1 GGMR01003864 MBY16483.1 AY060438 AAL25477.1 X17551 CAA35587.1 GFTR01008257 JAW08169.1 GGMR01013409 MBY26028.1 GEZM01084969 JAV60363.1 KU543673 AMS38352.1 DNOY01000259 HBE18313.1 AY508487 AAS13459.1 M38437 NEVH01005295 PNF38784.1 AY047531 AAK77263.1 GGMR01000226 MBY12845.1 ABLF02040174 ABLF02040183 ABLF02055266 GBHO01008295 JAG35309.1 GGMR01012443 MBY25062.1
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 KQ973342 KXZ75569.1 KQ971729 KYB24751.1 GALX01005359 JAB63107.1 GALX01005299 JAB63167.1 GBBI01004395 JAC14317.1 RPOV01000134 RPJ78669.1 GBBI01004571 JAC14141.1 KQ973412 EFA12510.2 S59870 AAB26437.2 GGMR01018716 MBY31335.1 ABLF02042518 ABLF02057746 GGMS01002992 MBY72195.1 GGFL01005012 MBW69190.1 GGFL01004957 MBW69135.1 GGMS01006359 MBY75562.1 ABLF02008153 ABLF02008462 ABLF02008464 GGFJ01001844 MBW50985.1 GGFJ01001842 MBW50983.1 GGFJ01001843 MBW50984.1 KK119692 KFM76444.1 GFTR01008038 JAW08388.1 NEVH01006736 PNF36716.1 GGFJ01001858 MBW50999.1 GGMR01003407 MBY16026.1 GGFJ01001857 MBW50998.1 NEVH01021205 PNF19912.1 GFTR01008271 JAW08155.1 GFTR01008279 JAW08147.1 GGMR01000676 MBY13295.1 NEVH01011985 PNF31054.1 GFTR01008261 JAW08165.1 GFTR01008256 JAW08170.1 NEVH01027074 PNF13841.1 GFTR01008251 JAW08175.1 NEVH01009134 PNF33528.1 GGMR01012305 MBY24924.1 GFTR01008262 JAW08164.1 GBGD01000415 JAC88474.1 NEVH01016302 PNF25823.1 ABLF02022418 GFXV01000908 MBW12713.1 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 ABLF02034467 GFTR01008269 JAW08157.1 ABLF02023911 NEVH01017470 PNF24199.1 ABLF02024246 GGMS01017365 MBY86568.1 GGFJ01001822 MBW50963.1 GGMR01006408 MBY19027.1 NEVH01020940 PNF20481.1 GFXV01002840 MBW14645.1 GGFL01005122 MBW69300.1 GGMR01016296 MBY28915.1 GGFL01005121 MBW69299.1 GFTR01008151 JAW08275.1 GBHO01010547 GBHO01010546 JAG33057.1 JAG33058.1 GFTR01008260 JAW08166.1 GGMR01012283 MBY24902.1 GBHO01021814 GBHO01010549 JAG21790.1 JAG33055.1 GGFJ01002256 MBW51397.1 GGMR01016905 MBY29524.1 GGFJ01002257 MBW51398.1 GEZM01053022 JAV74219.1 NEVH01026386 PNF14434.1 M22874 M38643 NWSH01007753 PCG62823.1 GFTR01008254 JAW08172.1 GBGD01000389 JAC88500.1 GGMR01003864 MBY16483.1 AY060438 AAL25477.1 X17551 CAA35587.1 GFTR01008257 JAW08169.1 GGMR01013409 MBY26028.1 GEZM01084969 JAV60363.1 KU543673 AMS38352.1 DNOY01000259 HBE18313.1 AY508487 AAS13459.1 M38437 NEVH01005295 PNF38784.1 AY047531 AAK77263.1 GGMR01000226 MBY12845.1 ABLF02040174 ABLF02040183 ABLF02055266 GBHO01008295 JAG35309.1 GGMR01012443 MBY25062.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
Q93137
Q9BPQ0
Q9XXW0
A0A0N1PFV1
O96546
A0A1Y1NA23
+ More
A0A139W8G9 A0A139WA11 V5GNF3 V5GNJ2 A0A023EYS8 A0A3N5Z7I3 A0A023EZE6 D7EM17 Q07995 A0A2S2PPK8 X1WLB2 A0A2S2Q383 A0A2M4CV42 A0A2M4CV52 A0A2S2QEE0 X1WNI2 A0A2M4BD66 A0A2M4BDA5 A0A2M4BD62 A0A087UGF5 A0A224XF24 A0A2J7R7D0 A0A2M4BD67 A0A2S2NGV8 A0A2M4BDV6 A0A2J7PU96 A0A224X6S1 A0A224X6Q4 A0A2S2N814 A0A2J7QR37 A0A224X6T4 A0A224XI99 A0A2J7PBY3 A0A224X6U5 A0A2J7QY69 A0A2S2P698 A0A224XB44 A0A069DZL8 A0A2J7QB68 X1WQI9 A0A2H8TF37 J9LC21 X1WZH0 A0A224X6R6 J9LFC9 A0A2J7Q6J2 J9M522 A0A2S2R9D4 A0A2M4BD83 A0A2S2NPG3 A0A2J7PVX3 A0A2H8TKI9 A0A2M4CVF1 A0A2S2PHL7 A0A2M4CVK2 A0A224X746 A0A0A9YJ00 A0A224X6W2 A0A2S2P6C4 A0A0A9YU04 A0A2M4BEF4 A0A2S2PJG2 A0A2M4BEZ9 A0A1Y1LKN6 A0A2J7PDM0 P21328 A0A2A4IUM2 A0A224XHW8 K7IN67 A0A069DX78 A0A2S2NH02 Q95SX7 Q04135 A0A224XJI0 A0A2S2PA10 A0A1Y1KFR3 A0A142LX26 A0A352X8G4 Q6R811 P21329 A0A2J7RD70 Q961V7 A0A2S2N6Q7 J9KLB1 A0A0A9YSX4 A0A2S2P6S3
A0A139W8G9 A0A139WA11 V5GNF3 V5GNJ2 A0A023EYS8 A0A3N5Z7I3 A0A023EZE6 D7EM17 Q07995 A0A2S2PPK8 X1WLB2 A0A2S2Q383 A0A2M4CV42 A0A2M4CV52 A0A2S2QEE0 X1WNI2 A0A2M4BD66 A0A2M4BDA5 A0A2M4BD62 A0A087UGF5 A0A224XF24 A0A2J7R7D0 A0A2M4BD67 A0A2S2NGV8 A0A2M4BDV6 A0A2J7PU96 A0A224X6S1 A0A224X6Q4 A0A2S2N814 A0A2J7QR37 A0A224X6T4 A0A224XI99 A0A2J7PBY3 A0A224X6U5 A0A2J7QY69 A0A2S2P698 A0A224XB44 A0A069DZL8 A0A2J7QB68 X1WQI9 A0A2H8TF37 J9LC21 X1WZH0 A0A224X6R6 J9LFC9 A0A2J7Q6J2 J9M522 A0A2S2R9D4 A0A2M4BD83 A0A2S2NPG3 A0A2J7PVX3 A0A2H8TKI9 A0A2M4CVF1 A0A2S2PHL7 A0A2M4CVK2 A0A224X746 A0A0A9YJ00 A0A224X6W2 A0A2S2P6C4 A0A0A9YU04 A0A2M4BEF4 A0A2S2PJG2 A0A2M4BEZ9 A0A1Y1LKN6 A0A2J7PDM0 P21328 A0A2A4IUM2 A0A224XHW8 K7IN67 A0A069DX78 A0A2S2NH02 Q95SX7 Q04135 A0A224XJI0 A0A2S2PA10 A0A1Y1KFR3 A0A142LX26 A0A352X8G4 Q6R811 P21329 A0A2J7RD70 Q961V7 A0A2S2N6Q7 J9KLB1 A0A0A9YSX4 A0A2S2P6S3
PDB
6AR3
E-value=0.0174653,
Score=93
Ontologies
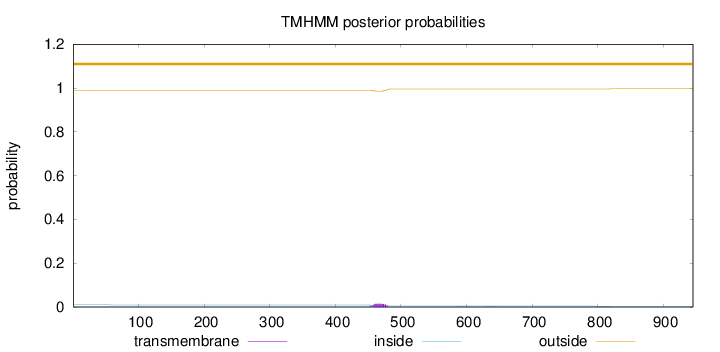
Topology
Length:
945
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.372630000000001
Exp number, first 60 AAs:
0.00127
Total prob of N-in:
0.00978
outside
1 - 945
Population Genetic Test Statistics
Pi
1.841369
Theta
14.318719
Tajima's D
0
CLR
22.911
CSRT
0.378331083445828
Interpretation
Uncertain
