Gene
KWMTBOMO14186 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011131
Annotation
NADH_dehydrogenase-ubiquinone_Fe-S_protein_2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.891 Mitochondrial Reliability : 2.28
Sequence
CDS
ATGGCCTCCCTTCTCAATGTTTTGTTGCGAAAATCTCCTAATTTCGGCCAAAAATCGGTCTTATTAAAGAACATTCCCGCTGTTCATAATGTGAACTCCCAAAGAGATGGGCACCGTTGGTTCCCTGATCCAGACTTTGTGAAACAATTTGAAGGGCCAGTCATGTATCCTGATGAGTCGACCAAATCATTGAAACCTGTTCCTTATAACAGCATCATAAAACCGGCCGAAAAGAAAGTACGCAACATGATCCTGAACTTCGGGCCCCAGCATCCAGCCGCTCACGGGGTGCTGCGTCTCGTACTGGAACTGGACGGAGAGACAGTCCGTGCAGCAGATCCTCACATAGGTCTCCTGCACCGTGGTACTGAAAAACTGATCGAATACAAGACATACACCCAAGCCCTGCCGTACTTCGACCGTCTGGACTACGTGTCCATGATGTGCAACGAGCAGTGCTACAGTCTGGCCGTCGAGAAGCTGCTGAACATCGATGTTCCTCTGCGGGCCAAGTACATCAGAACTCTGTTTGCGGAGATAACCCGCCTACTGAACCACATCATGGCGGTCGGCACGCACGCGCTCGACGTGGGGGCGCTCACTCCCTTCTTCTGGCTGTTTGAGGAGAGAGAGAAGATGATGGAGTTCTACGAGAGAGTCAGCGGCGCCAGGATGCACGCCGCCTACATCAGACCAGGAGGGGTTTCGCTTGACATGCCGCTGGGTCTGATGGACGACATCTACGAGTTCTCGTCGAAGTTCGCCGAGCGCATCGACGAGGTGGAGGACGTGCTCACCACCAACAGGATCTGGGTGCAGCGGACCAAGGACATCGGCGTGGTCACTGCGCAGGACGCGCTCAACTACGGCTTCAGCGGTGTGATGCTGCGGGGCTCCGGCATCAAGTGGGACCTCCGCAAGACGCAGCCCTACGACGCCTATCCCTTCGTCGAGTTCGACGTGCCCATAGGGACGCACGGCGACTGCTACGATAGGTACTTGTGTCGCGTCGAGGAGATGCGCCAGTCCCTCCGCATCATCGAGCAGTGCCTCAACGACATGCCCCCCGGGGAGGTCAAGACCGACGACGCCAAGCTCACCCCGCCCTCCCGGGAGGAGATGAAGACGTCCATGGAGGCGCTGATCCACCACTTCAAGCTGTTCACGCAGGGCTACCAGGTGCCGCCGGGCTCCACCTACACCGCCGTGGAGGCTCCCAAGGGGGAGTTCGGGATCTACCTCGTCTCCGACGGAGGCTCCAAGCCGTACAGATGCAAGATCAAGGCGCCGGGCTTCGCGCACCTGGCGGCGCTGGAGAAGATCGGCAGGAACTCCATGCTGGCGGACATCGTGGCCATCATCGGGACCCTCGACGTGGTCTTCGGGGAGATCGACCGGTAG
Protein
MASLLNVLLRKSPNFGQKSVLLKNIPAVHNVNSQRDGHRWFPDPDFVKQFEGPVMYPDESTKSLKPVPYNSIIKPAEKKVRNMILNFGPQHPAAHGVLRLVLELDGETVRAADPHIGLLHRGTEKLIEYKTYTQALPYFDRLDYVSMMCNEQCYSLAVEKLLNIDVPLRAKYIRTLFAEITRLLNHIMAVGTHALDVGALTPFFWLFEEREKMMEFYERVSGARMHAAYIRPGGVSLDMPLGLMDDIYEFSSKFAERIDEVEDVLTTNRIWVQRTKDIGVVTAQDALNYGFSGVMLRGSGIKWDLRKTQPYDAYPFVEFDVPIGTHGDCYDRYLCRVEEMRQSLRIIEQCLNDMPPGEVKTDDAKLTPPSREEMKTSMEALIHHFKLFTQGYQVPPGSTYTAVEAPKGEFGIYLVSDGGSKPYRCKIKAPGFAHLAALEKIGRNSMLADIVAIIGTLDVVFGEIDR
Summary
Similarity
Belongs to the complex I 49 kDa subunit family.
Uniprot
Q1HQ80
A0A2W1BJF4
A0A2A4IYD3
A0A3S2LWJ9
W8S0Q1
S4NXW7
+ More
A0A194PSU9 A0A212FJU2 H9JNM7 W8BPS0 A0A034WIL4 A0A0K8U9R6 A0A2M4A5Q1 A0A084WPL7 A0A182HFH3 A0A1W4V5B6 A0A182JHJ2 B4JZW3 A0A2M4A5R4 I4DNK1 D2A579 B3P9S8 A0A182FIL8 Q16Q68 B4NHW8 A0A2M3Z2M7 A0A2M3Z2Q1 Q9V4E0 A0A2M4BM20 T1E9X3 B5DTD6 A0A3B0KUM6 U5EVJ5 B4L7B6 B4PW13 A0A182PM65 B4IIX1 A0A0J9S0T2 B4H874 A0A182LZ26 A0A182XY39 A0A182I9M2 A0A182X5B3 A0A0M4ETC2 A0A182TTJ2 A0A1J1HIR6 A0A182UZE7 T1PH29 W5JI89 A0A182N0Z0 A0A182QSH7 A0A1Q3FM55 A0A0K8TTI9 B0WWS9 B4MF01 A0A182KFB3 A0A1I8P7E3 A0A1L8EJC6 B3N044 A0A182IKS4 F5HJY7 A0A1B0CDK6 A0A182RJ19 A0A182VVB4 A0A182MPT1 A0A3B0K8N8 A0A1L8E0G2 V5GWA2 A0A1A9WEW4 A0A1L8E0L2 A0A182WBD4 J3JUS9 A0A084VY01 A0A182K1D4 A0A182PIN9 A0A1W4X418 A0A1B0BFM9 A0A1A9ZL17 A0A1A9XFD2 D3TR12 A0A182V7Y6 A0A336M652 A0A182I6L0 A0A182L3U2 A0A1A9V8L5 Q7PTG6 A0A1Y1MKB5 A0A182WWD1 N6TVB5 A0A023EU66 A0A1B6C2M5 A0A0T6AX38 A0A182Y825 A0A182UDY9 U4U242 A0A1D2NDE4 A0A023FA96 A0A182LD64 A0A224XC61 A0A146LBH5
A0A194PSU9 A0A212FJU2 H9JNM7 W8BPS0 A0A034WIL4 A0A0K8U9R6 A0A2M4A5Q1 A0A084WPL7 A0A182HFH3 A0A1W4V5B6 A0A182JHJ2 B4JZW3 A0A2M4A5R4 I4DNK1 D2A579 B3P9S8 A0A182FIL8 Q16Q68 B4NHW8 A0A2M3Z2M7 A0A2M3Z2Q1 Q9V4E0 A0A2M4BM20 T1E9X3 B5DTD6 A0A3B0KUM6 U5EVJ5 B4L7B6 B4PW13 A0A182PM65 B4IIX1 A0A0J9S0T2 B4H874 A0A182LZ26 A0A182XY39 A0A182I9M2 A0A182X5B3 A0A0M4ETC2 A0A182TTJ2 A0A1J1HIR6 A0A182UZE7 T1PH29 W5JI89 A0A182N0Z0 A0A182QSH7 A0A1Q3FM55 A0A0K8TTI9 B0WWS9 B4MF01 A0A182KFB3 A0A1I8P7E3 A0A1L8EJC6 B3N044 A0A182IKS4 F5HJY7 A0A1B0CDK6 A0A182RJ19 A0A182VVB4 A0A182MPT1 A0A3B0K8N8 A0A1L8E0G2 V5GWA2 A0A1A9WEW4 A0A1L8E0L2 A0A182WBD4 J3JUS9 A0A084VY01 A0A182K1D4 A0A182PIN9 A0A1W4X418 A0A1B0BFM9 A0A1A9ZL17 A0A1A9XFD2 D3TR12 A0A182V7Y6 A0A336M652 A0A182I6L0 A0A182L3U2 A0A1A9V8L5 Q7PTG6 A0A1Y1MKB5 A0A182WWD1 N6TVB5 A0A023EU66 A0A1B6C2M5 A0A0T6AX38 A0A182Y825 A0A182UDY9 U4U242 A0A1D2NDE4 A0A023FA96 A0A182LD64 A0A224XC61 A0A146LBH5
Pubmed
28756777
23622113
26354079
22118469
19121390
24495485
+ More
25348373 24438588 26483478 17994087 22651552 18362917 19820115 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 22936249 25244985 25315136 20920257 23761445 26369729 18057021 20479145 12364791 14747013 17210077 22516182 20353571 20966253 28004739 23537049 24945155 27289101 25474469 26823975
25348373 24438588 26483478 17994087 22651552 18362917 19820115 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 22936249 25244985 25315136 20920257 23761445 26369729 18057021 20479145 12364791 14747013 17210077 22516182 20353571 20966253 28004739 23537049 24945155 27289101 25474469 26823975
EMBL
DQ443172
ABF51261.1
KZ150147
PZC72870.1
NWSH01004637
PCG64835.1
+ More
RSAL01000156 RVE45606.1 KF994920 AHL83459.1 GAIX01011992 JAA80568.1 KQ459593 KPI96387.1 AGBW02008219 OWR53996.1 BABH01035080 GAMC01005613 JAC00943.1 GAKP01004790 JAC54162.1 GDHF01029219 JAI23095.1 GGFK01002806 MBW36127.1 ATLV01025101 KE525369 KFB52161.1 JXUM01036889 KQ561109 KXJ79679.1 CH916380 EDV90979.1 GGFK01002798 MBW36119.1 AK402977 BAM19491.1 KQ971345 EFA05697.1 CH954184 EDV45241.1 CH477759 EAT36538.1 CH964272 EDW83618.1 GGFM01002004 MBW22755.1 GGFM01002048 MBW22799.1 AE014135 BT032847 AAF59336.2 ACD81861.1 AGB96554.1 GGFJ01004850 MBW53991.1 GAMD01001523 JAB00068.1 CH475843 EDY71141.2 OUUW01000017 SPP89021.1 GANO01001832 JAB58039.1 CH933813 EDW10910.2 CM000161 EDW99318.1 CH480845 EDW50912.1 CM002916 KMZ07247.1 CH479221 EDW34874.1 AXCM01009391 APCN01001005 CP012527 ALC48135.1 CVRI01000004 CRK87450.1 KA647440 AFP62069.1 ADMH02001172 ETN63841.1 AXCN02001373 GFDL01006441 JAV28604.1 GDAI01000132 JAI17471.1 DS232152 EDS36190.1 CM000831 CH940665 ACY70475.1 EDW71102.1 GFDG01000045 JAV18754.1 CH902639 EDV33552.1 AAAB01008847 EAA06952.3 EGK96602.1 EGK96603.1 AJWK01007963 AXCM01009889 SPP89022.1 GFDF01001866 JAV12218.1 GALX01003958 JAB64508.1 GFDF01001865 JAV12219.1 BT126994 AEE61956.1 ATLV01018235 KE525226 KFB42845.1 JXJN01013592 CCAG010006472 EZ423864 ADD20140.1 UFQT01000445 SSX24369.1 APCN01003658 AAAB01008807 EAA03959.4 GEZM01028984 JAV86171.1 APGK01050477 APGK01050478 KB741174 ENN73220.1 GAPW01001644 JAC11954.1 GEDC01029535 JAS07763.1 LJIG01022656 KRT79413.1 KB631091 KB631092 ERL84090.1 ERL84096.1 LJIJ01000079 ODN03287.1 GBBI01000869 JAC17843.1 GFTR01006449 JAW09977.1 GDHC01014093 JAQ04536.1
RSAL01000156 RVE45606.1 KF994920 AHL83459.1 GAIX01011992 JAA80568.1 KQ459593 KPI96387.1 AGBW02008219 OWR53996.1 BABH01035080 GAMC01005613 JAC00943.1 GAKP01004790 JAC54162.1 GDHF01029219 JAI23095.1 GGFK01002806 MBW36127.1 ATLV01025101 KE525369 KFB52161.1 JXUM01036889 KQ561109 KXJ79679.1 CH916380 EDV90979.1 GGFK01002798 MBW36119.1 AK402977 BAM19491.1 KQ971345 EFA05697.1 CH954184 EDV45241.1 CH477759 EAT36538.1 CH964272 EDW83618.1 GGFM01002004 MBW22755.1 GGFM01002048 MBW22799.1 AE014135 BT032847 AAF59336.2 ACD81861.1 AGB96554.1 GGFJ01004850 MBW53991.1 GAMD01001523 JAB00068.1 CH475843 EDY71141.2 OUUW01000017 SPP89021.1 GANO01001832 JAB58039.1 CH933813 EDW10910.2 CM000161 EDW99318.1 CH480845 EDW50912.1 CM002916 KMZ07247.1 CH479221 EDW34874.1 AXCM01009391 APCN01001005 CP012527 ALC48135.1 CVRI01000004 CRK87450.1 KA647440 AFP62069.1 ADMH02001172 ETN63841.1 AXCN02001373 GFDL01006441 JAV28604.1 GDAI01000132 JAI17471.1 DS232152 EDS36190.1 CM000831 CH940665 ACY70475.1 EDW71102.1 GFDG01000045 JAV18754.1 CH902639 EDV33552.1 AAAB01008847 EAA06952.3 EGK96602.1 EGK96603.1 AJWK01007963 AXCM01009889 SPP89022.1 GFDF01001866 JAV12218.1 GALX01003958 JAB64508.1 GFDF01001865 JAV12219.1 BT126994 AEE61956.1 ATLV01018235 KE525226 KFB42845.1 JXJN01013592 CCAG010006472 EZ423864 ADD20140.1 UFQT01000445 SSX24369.1 APCN01003658 AAAB01008807 EAA03959.4 GEZM01028984 JAV86171.1 APGK01050477 APGK01050478 KB741174 ENN73220.1 GAPW01001644 JAC11954.1 GEDC01029535 JAS07763.1 LJIG01022656 KRT79413.1 KB631091 KB631092 ERL84090.1 ERL84096.1 LJIJ01000079 ODN03287.1 GBBI01000869 JAC17843.1 GFTR01006449 JAW09977.1 GDHC01014093 JAQ04536.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007151
UP000005204
UP000030765
+ More
UP000069940 UP000249989 UP000192221 UP000075880 UP000001070 UP000007266 UP000008711 UP000069272 UP000008820 UP000007798 UP000000803 UP000001819 UP000268350 UP000009192 UP000002282 UP000075885 UP000001292 UP000008744 UP000075883 UP000076408 UP000075840 UP000076407 UP000092553 UP000075902 UP000183832 UP000075903 UP000095301 UP000000673 UP000075884 UP000075886 UP000002320 UP000008792 UP000075881 UP000095300 UP000007801 UP000007062 UP000092461 UP000075900 UP000075920 UP000091820 UP000192223 UP000092460 UP000092445 UP000092443 UP000092444 UP000075882 UP000078200 UP000019118 UP000030742 UP000094527
UP000069940 UP000249989 UP000192221 UP000075880 UP000001070 UP000007266 UP000008711 UP000069272 UP000008820 UP000007798 UP000000803 UP000001819 UP000268350 UP000009192 UP000002282 UP000075885 UP000001292 UP000008744 UP000075883 UP000076408 UP000075840 UP000076407 UP000092553 UP000075902 UP000183832 UP000075903 UP000095301 UP000000673 UP000075884 UP000075886 UP000002320 UP000008792 UP000075881 UP000095300 UP000007801 UP000007062 UP000092461 UP000075900 UP000075920 UP000091820 UP000192223 UP000092460 UP000092445 UP000092443 UP000092444 UP000075882 UP000078200 UP000019118 UP000030742 UP000094527
Interpro
SUPFAM
SSF56762
SSF56762
Gene 3D
ProteinModelPortal
Q1HQ80
A0A2W1BJF4
A0A2A4IYD3
A0A3S2LWJ9
W8S0Q1
S4NXW7
+ More
A0A194PSU9 A0A212FJU2 H9JNM7 W8BPS0 A0A034WIL4 A0A0K8U9R6 A0A2M4A5Q1 A0A084WPL7 A0A182HFH3 A0A1W4V5B6 A0A182JHJ2 B4JZW3 A0A2M4A5R4 I4DNK1 D2A579 B3P9S8 A0A182FIL8 Q16Q68 B4NHW8 A0A2M3Z2M7 A0A2M3Z2Q1 Q9V4E0 A0A2M4BM20 T1E9X3 B5DTD6 A0A3B0KUM6 U5EVJ5 B4L7B6 B4PW13 A0A182PM65 B4IIX1 A0A0J9S0T2 B4H874 A0A182LZ26 A0A182XY39 A0A182I9M2 A0A182X5B3 A0A0M4ETC2 A0A182TTJ2 A0A1J1HIR6 A0A182UZE7 T1PH29 W5JI89 A0A182N0Z0 A0A182QSH7 A0A1Q3FM55 A0A0K8TTI9 B0WWS9 B4MF01 A0A182KFB3 A0A1I8P7E3 A0A1L8EJC6 B3N044 A0A182IKS4 F5HJY7 A0A1B0CDK6 A0A182RJ19 A0A182VVB4 A0A182MPT1 A0A3B0K8N8 A0A1L8E0G2 V5GWA2 A0A1A9WEW4 A0A1L8E0L2 A0A182WBD4 J3JUS9 A0A084VY01 A0A182K1D4 A0A182PIN9 A0A1W4X418 A0A1B0BFM9 A0A1A9ZL17 A0A1A9XFD2 D3TR12 A0A182V7Y6 A0A336M652 A0A182I6L0 A0A182L3U2 A0A1A9V8L5 Q7PTG6 A0A1Y1MKB5 A0A182WWD1 N6TVB5 A0A023EU66 A0A1B6C2M5 A0A0T6AX38 A0A182Y825 A0A182UDY9 U4U242 A0A1D2NDE4 A0A023FA96 A0A182LD64 A0A224XC61 A0A146LBH5
A0A194PSU9 A0A212FJU2 H9JNM7 W8BPS0 A0A034WIL4 A0A0K8U9R6 A0A2M4A5Q1 A0A084WPL7 A0A182HFH3 A0A1W4V5B6 A0A182JHJ2 B4JZW3 A0A2M4A5R4 I4DNK1 D2A579 B3P9S8 A0A182FIL8 Q16Q68 B4NHW8 A0A2M3Z2M7 A0A2M3Z2Q1 Q9V4E0 A0A2M4BM20 T1E9X3 B5DTD6 A0A3B0KUM6 U5EVJ5 B4L7B6 B4PW13 A0A182PM65 B4IIX1 A0A0J9S0T2 B4H874 A0A182LZ26 A0A182XY39 A0A182I9M2 A0A182X5B3 A0A0M4ETC2 A0A182TTJ2 A0A1J1HIR6 A0A182UZE7 T1PH29 W5JI89 A0A182N0Z0 A0A182QSH7 A0A1Q3FM55 A0A0K8TTI9 B0WWS9 B4MF01 A0A182KFB3 A0A1I8P7E3 A0A1L8EJC6 B3N044 A0A182IKS4 F5HJY7 A0A1B0CDK6 A0A182RJ19 A0A182VVB4 A0A182MPT1 A0A3B0K8N8 A0A1L8E0G2 V5GWA2 A0A1A9WEW4 A0A1L8E0L2 A0A182WBD4 J3JUS9 A0A084VY01 A0A182K1D4 A0A182PIN9 A0A1W4X418 A0A1B0BFM9 A0A1A9ZL17 A0A1A9XFD2 D3TR12 A0A182V7Y6 A0A336M652 A0A182I6L0 A0A182L3U2 A0A1A9V8L5 Q7PTG6 A0A1Y1MKB5 A0A182WWD1 N6TVB5 A0A023EU66 A0A1B6C2M5 A0A0T6AX38 A0A182Y825 A0A182UDY9 U4U242 A0A1D2NDE4 A0A023FA96 A0A182LD64 A0A224XC61 A0A146LBH5
PDB
6G72
E-value=0,
Score=1887
Ontologies
PATHWAY
GO
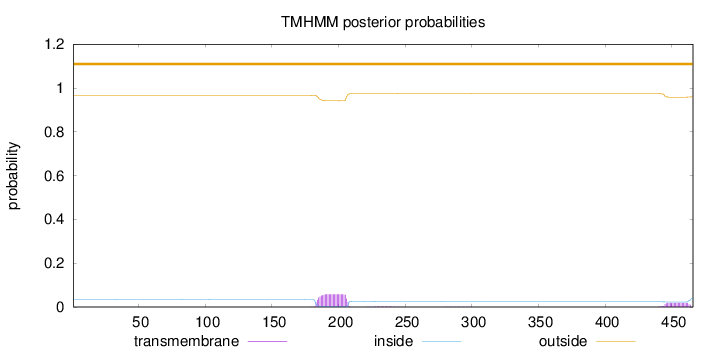
Topology
Length:
466
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.69417
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03345
outside
1 - 466
Population Genetic Test Statistics
Pi
15.749772
Theta
15.884976
Tajima's D
-1.010105
CLR
0.835579
CSRT
0.133393330333483
Interpretation
Uncertain
