Gene
KWMTBOMO14185 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011133
Annotation
PREDICTED:_protein_suppressor_of_forked_[Papilio_xuthus]
Full name
Protein suppressor of forked
Location in the cell
Cytoplasmic Reliability : 2.237
Sequence
CDS
ATGGCATCTGTAGATGAAATCGCCGACATTGATTGGGGTAATGAACGCCTATCCCGCGCCCAGCGTGCAGTCGAAGCCAACATATACGATGTGGACTCTTGGTCATTGCTAGTAAGAGAAGCCCAGGCCCGAAACATCAACGATGTCCGAACCATGTACGAGAAGTTGATAACGGCATTCCCGACCACAGGACGTTACTGGAAAATATATATCGAACAGGAGATGAAAGCGAGAAATTTTGAAAGAGTCGAAAAGCTCTTCCAGAGGTGTCTGATGAAGATATTAAACATCGAACTCTGGAGGCTGTACCTGAACTACGTGAAGGAGACGAAGTGCATGCTACCCACTTACAAGGAGAAAATGGCGCAGGCCTACGACTTCGCCCTGGACAAGATAGGTCTGGACATCCACGCTTACCCAATATGGAACGACTACGTGACGTTTCTGAAGAGTGTCGAAGCCGTTGGTTCATACGCCGAGAACCAGAAGATATCCGCCGTTCGAAAGATCTATCAGAGAGCGGTCATAACGCCCATAATAGGCATAGAGACGCTGTGGAAAGACTATATAGCCTTCGAGCAAAGCATCAACCCGATAATCGCCGAAAGGATGGCCATGGAGCGTTCCCGGGAGTACATGAACGCGAGACGAGTCTCCAAGGAGCTGGAGACGGTCACCCGGGGACTCAATAGGAACATGCCCGCGACGCCGCCCACCGTCGACCGCGAGGAGATGAAACAGGTGGAGTTATGGAAGAAGTACATATCGTGGGAGCGCGCGAACCCCCTCCGCTCCGAGGACACGGCGCTGGTGGCGCGGCGCGTGATGTTCGCCATCGAGCAGTGCCTGCTGTGTCTGGCGCACCACCCGGACGTGTGGCACCAGGCCGCGCAGTTCCTCGACCACTCCTCCAAGCTGCTGCAGGAGAAGGGGGACTCGAACGCGTCCCGGCTGTTCTCTGACGAGGCGGCGGCGGTGTACGAGCGCGCCACCAGCGGCCCCCTCAAACACTCCACGCTGCTGCACTTCGCACACGCAGACTACGAGGAGAGCAGGCTGCACTACCACAAGGTGCACCAGGTCTACACGCGCTACCTGGACACAGAGGACATCGACCCCACGTTGGCCTATGTACAGTACATGAAGTTCGCCAGGAGGGCGGAGGGAATCAAGTCTGCGAGGACCGTCTTCAAGAGGGCCCGGGAGGATCAGAGGTCCCGGTACCACGTGTTCGTGGCCGCCGCGCTCATGGAGTACTACTGCTCCAAGGACAAGAACATAGCCTTCAGGATCTTCGAGCTCGGCCTCAAGAAGTTCTCCCACATCCCGGAGTACGTGCTCTGCTACATCGACTACCTGTCCCATCTCAACGAGGATAACAACACGAGGGTTCTGTTCGAGCGGGTGCTGTCCTCGGGCAGCCTGAGGCCGGAGCACTCAGTCGACATATGGAACAGATTCCTGGAGTTCGAGTCCAACATCGGGGACCTGGTCAGCATTGTCAAAGTCGAGAAGCGGCGGCAGGGCGTGCTCGAGAAGATAAAAGAATTCGAAGGCAAAGAGACGGCCCAGCTGGTGGACAGATACAAGTTCCTCGACCTGTACCCGTGCAGCATCGCCGAGCTGAAGTCTATAGGGTACACTGAGGTGGCCTCCATGTCGAACAAGTCGTGGGCCCTCGGGGGGCCGCTGGCGGGGATCTCGCCGGAGCTGGCGGCCGTCATCCTGGGGCAGAGAGACAACGATCCTAACAAAGACATTGCGCGTCCGGACACCAACCAAATGATTCCGTACAAACCTAAGTCGAACCCACTACCCGGGGAGCACCCCATACCAGGTGGGGCGTTTCCGCTGCCGTCGAGCGCGGCGCACGTGTGCACGCTGCTGCCGCCGCCCAACTGCTTCCGGGGACCCTTCGTGGCGGTGGACCGGCTCATGCAGCTCTTCAACAGGATATCGCTCCCGGATAAAGTGCCGGCGCCCTCCGCGGAGAACGGCTGCGACACGCGGCTGTTCGAGATGGCGCGCTCCGTGCACTGGCTGTTCGACGACGACGGCGTCACCGCCATCTCCAATAAGGTCCGGCGCAGGAAGCTGGGCGACGACTCTGACGACGACGAGCTGGGCACGGCGCCCCCGGTCAACGACATATACAGGCAGCGGCAGCAGAAGCGCGTCAAGTAG
Protein
MASVDEIADIDWGNERLSRAQRAVEANIYDVDSWSLLVREAQARNINDVRTMYEKLITAFPTTGRYWKIYIEQEMKARNFERVEKLFQRCLMKILNIELWRLYLNYVKETKCMLPTYKEKMAQAYDFALDKIGLDIHAYPIWNDYVTFLKSVEAVGSYAENQKISAVRKIYQRAVITPIIGIETLWKDYIAFEQSINPIIAERMAMERSREYMNARRVSKELETVTRGLNRNMPATPPTVDREEMKQVELWKKYISWERANPLRSEDTALVARRVMFAIEQCLLCLAHHPDVWHQAAQFLDHSSKLLQEKGDSNASRLFSDEAAAVYERATSGPLKHSTLLHFAHADYEESRLHYHKVHQVYTRYLDTEDIDPTLAYVQYMKFARRAEGIKSARTVFKRAREDQRSRYHVFVAAALMEYYCSKDKNIAFRIFELGLKKFSHIPEYVLCYIDYLSHLNEDNNTRVLFERVLSSGSLRPEHSVDIWNRFLEFESNIGDLVSIVKVEKRRQGVLEKIKEFEGKETAQLVDRYKFLDLYPCSIAELKSIGYTEVASMSNKSWALGGPLAGISPELAAVILGQRDNDPNKDIARPDTNQMIPYKPKSNPLPGEHPIPGGAFPLPSSAAHVCTLLPPPNCFRGPFVAVDRLMQLFNRISLPDKVPAPSAENGCDTRLFEMARSVHWLFDDDGVTAISNKVRRRKLGDDSDDDELGTAPPVNDIYRQRQQKRVK
Summary
Description
Essential protein, may play a role in mRNA production or stability.
Subunit
Probably interacts with an RNA-binding protein.
Similarity
Belongs to the Ca(2+):cation antiporter (CaCA) (TC 2.A.19) family.
Keywords
Alternative splicing
Complete proteome
Nucleus
Phosphoprotein
Reference proteome
Repeat
Feature
chain Protein suppressor of forked
splice variant In isoform E.
splice variant In isoform E.
Uniprot
H9JNM9
A0A2A4IZY6
A0A194PSH1
A0A2W1BC53
A0A212FJS9
A0A2H1WFW9
+ More
A0A194RMW6 A0A154PG94 A0A0C9S0B0 A0A0M8ZTK3 A0A310S893 A0A088AKK0 A0A0L7R9J2 E2AWC7 A0A067R195 A0A232ENU7 A0A026X3Y1 A0A3L8DG90 A0A195AZG7 A0A195EE00 F4X1W4 E2BXK4 A0A195D4K0 A0A1B6CGC0 T1HT88 A0A1W4X9X1 A0A023F598 A0A2A3EH94 A0A0J7KXU5 A0A1B6KX04 A0A195FYA3 D2A3P0 A0A0A9YM07 N6SVB2 A0A1B6K0U8 A0A1B6ENK3 A0A182Y4B2 A0A182JTF5 A0A182P5T0 A0A182IRV9 A0A182RNQ7 A0A182QHX7 A0A1B0DCA6 A0A1B6J9Z5 A0A084VRC4 A0A2M4AMV5 A0A182F7E1 A0A1B0CCL9 A0A2M4BF68 A0A151X8E4 A0A182MUI8 A0A1B6GER8 W5JIY1 A0A182VE99 A0A182WRJ2 Q7QDA0 A0A182HU85 A0A182TDN5 A0A182N635 A0A1S4FBE7 A0A0K8VUG6 A0A034VYT8 A0A1L8DGX8 A0A1I8M8R5 Q0IFE2 A0A1A9W3Z9 A0A1J1HRE8 A0A1B0AC18 A0A0L0BYQ7 A0A1A9XE12 A0A1I8M8S0 A0A1I8PF42 A0A1Q3G0R8 A0A1B0BVY6 E0VSV5 A0A2M4ANX4 B4L585 B3NYA3 A0A0J9R6P7 J9JQD1 P25991-4 A0A0P8XS09 A0A0R1E736 B4IM34 A0A1W4WDD0 B4NEU6 A0A0M5J1D0 A0A158NHM7 A0A2S2R309 B5DLV3 A0A2M4BGT1 A0A1W7R5N0 Q6V8Y2 A0A3B0K687 O96641 B4JKH5 B4H4H0 A0A182W069 B0W9I8 A0A226ETM0
A0A194RMW6 A0A154PG94 A0A0C9S0B0 A0A0M8ZTK3 A0A310S893 A0A088AKK0 A0A0L7R9J2 E2AWC7 A0A067R195 A0A232ENU7 A0A026X3Y1 A0A3L8DG90 A0A195AZG7 A0A195EE00 F4X1W4 E2BXK4 A0A195D4K0 A0A1B6CGC0 T1HT88 A0A1W4X9X1 A0A023F598 A0A2A3EH94 A0A0J7KXU5 A0A1B6KX04 A0A195FYA3 D2A3P0 A0A0A9YM07 N6SVB2 A0A1B6K0U8 A0A1B6ENK3 A0A182Y4B2 A0A182JTF5 A0A182P5T0 A0A182IRV9 A0A182RNQ7 A0A182QHX7 A0A1B0DCA6 A0A1B6J9Z5 A0A084VRC4 A0A2M4AMV5 A0A182F7E1 A0A1B0CCL9 A0A2M4BF68 A0A151X8E4 A0A182MUI8 A0A1B6GER8 W5JIY1 A0A182VE99 A0A182WRJ2 Q7QDA0 A0A182HU85 A0A182TDN5 A0A182N635 A0A1S4FBE7 A0A0K8VUG6 A0A034VYT8 A0A1L8DGX8 A0A1I8M8R5 Q0IFE2 A0A1A9W3Z9 A0A1J1HRE8 A0A1B0AC18 A0A0L0BYQ7 A0A1A9XE12 A0A1I8M8S0 A0A1I8PF42 A0A1Q3G0R8 A0A1B0BVY6 E0VSV5 A0A2M4ANX4 B4L585 B3NYA3 A0A0J9R6P7 J9JQD1 P25991-4 A0A0P8XS09 A0A0R1E736 B4IM34 A0A1W4WDD0 B4NEU6 A0A0M5J1D0 A0A158NHM7 A0A2S2R309 B5DLV3 A0A2M4BGT1 A0A1W7R5N0 Q6V8Y2 A0A3B0K687 O96641 B4JKH5 B4H4H0 A0A182W069 B0W9I8 A0A226ETM0
Pubmed
19121390
26354079
28756777
22118469
20798317
24845553
+ More
28648823 24508170 30249741 21719571 25474469 18362917 19820115 25401762 26823975 23537049 25244985 24438588 20920257 23761445 12364791 14747013 17210077 25348373 25315136 17510324 26108605 20566863 17994087 22936249 8436295 10731132 12537572 12537569 17372656 18327897 17550304 21347285 15632085 9826695 18057021
28648823 24508170 30249741 21719571 25474469 18362917 19820115 25401762 26823975 23537049 25244985 24438588 20920257 23761445 12364791 14747013 17210077 25348373 25315136 17510324 26108605 20566863 17994087 22936249 8436295 10731132 12537572 12537569 17372656 18327897 17550304 21347285 15632085 9826695 18057021
EMBL
BABH01035077
BABH01035078
NWSH01004637
PCG64834.1
KQ459593
KPI96386.1
+ More
KZ150147 PZC72869.1 AGBW02008219 OWR53995.1 ODYU01008301 SOQ51772.1 KQ460152 KPJ17341.1 KQ434889 KZC10328.1 GBYB01015250 JAG85017.1 KQ435894 KOX69353.1 KQ764772 OAD54401.1 KQ414624 KOC67537.1 GL443286 EFN62227.1 KK852785 KDR16574.1 NNAY01003069 OXU20030.1 KK107031 EZA62089.1 QOIP01000008 RLU19326.1 KQ976694 KYM77633.1 KQ979039 KYN23433.1 GL888555 EGI59551.1 GL451265 EFN79573.1 KQ976870 KYN07813.1 GEDC01024752 GEDC01007151 JAS12546.1 JAS30147.1 ACPB03006075 GBBI01002439 JAC16273.1 KZ288248 PBC31163.1 LBMM01002026 KMQ95377.1 GEBQ01024017 JAT15960.1 KQ981208 KYN44814.1 KQ971348 EFA05540.1 GBHO01011496 GBRD01007570 GDHC01001156 JAG32108.1 JAG58251.1 JAQ17473.1 APGK01055258 KB741261 KB632413 ENN71639.1 ERL95267.1 GECU01002658 JAT05049.1 GECZ01030281 JAS39488.1 AXCN02000193 AJVK01030700 GECU01011706 JAS96000.1 ATLV01015604 KE525023 KFB40518.1 GGFK01008790 MBW42111.1 AJWK01006903 GGFJ01002559 MBW51700.1 KQ982409 KYQ56647.1 AXCM01000884 GECZ01008841 JAS60928.1 ADMH02001371 ETN62749.1 AAAB01008859 EAA07937.5 APCN01000612 GDHF01032141 GDHF01015087 GDHF01009790 GDHF01003337 JAI20173.1 JAI37227.1 JAI42524.1 JAI48977.1 GAKP01011728 GAKP01011727 GAKP01011726 GAKP01011725 JAC47227.1 GFDF01008447 JAV05637.1 CH477356 EAT42772.1 CVRI01000020 CRK90547.1 JRES01001143 KNC25160.1 GFDL01001655 JAV33390.1 JXJN01021534 DS235758 EEB16461.1 GGFK01009152 MBW42473.1 CH933811 EDW06344.1 CH954180 EDV47688.1 CM002911 KMY91726.1 ABLF02031355 X62679 AE014298 BT012671 BT021384 AY102664 CH902630 KPU77416.1 CH891579 KRK05040.1 CH480924 EDW43670.1 CH964239 EDW82265.1 CP012528 ALC49674.1 ADTU01015668 ADTU01015669 ADTU01015670 ADTU01015671 ADTU01015672 ADTU01015673 ADTU01015674 ADTU01015675 ADTU01015676 GGMS01014519 MBY83722.1 CH379064 EDY72500.2 GGFJ01003020 MBW52161.1 GEHC01001212 JAV46433.1 AY347747 AAR01873.1 OUUW01000025 SPP89644.1 AF097830 CH940653 AAC99436.1 KRF80528.1 CH916370 EDW00078.1 CH479209 EDW32721.1 DS231864 EDS40213.1 LNIX01000002 OXA60580.1
KZ150147 PZC72869.1 AGBW02008219 OWR53995.1 ODYU01008301 SOQ51772.1 KQ460152 KPJ17341.1 KQ434889 KZC10328.1 GBYB01015250 JAG85017.1 KQ435894 KOX69353.1 KQ764772 OAD54401.1 KQ414624 KOC67537.1 GL443286 EFN62227.1 KK852785 KDR16574.1 NNAY01003069 OXU20030.1 KK107031 EZA62089.1 QOIP01000008 RLU19326.1 KQ976694 KYM77633.1 KQ979039 KYN23433.1 GL888555 EGI59551.1 GL451265 EFN79573.1 KQ976870 KYN07813.1 GEDC01024752 GEDC01007151 JAS12546.1 JAS30147.1 ACPB03006075 GBBI01002439 JAC16273.1 KZ288248 PBC31163.1 LBMM01002026 KMQ95377.1 GEBQ01024017 JAT15960.1 KQ981208 KYN44814.1 KQ971348 EFA05540.1 GBHO01011496 GBRD01007570 GDHC01001156 JAG32108.1 JAG58251.1 JAQ17473.1 APGK01055258 KB741261 KB632413 ENN71639.1 ERL95267.1 GECU01002658 JAT05049.1 GECZ01030281 JAS39488.1 AXCN02000193 AJVK01030700 GECU01011706 JAS96000.1 ATLV01015604 KE525023 KFB40518.1 GGFK01008790 MBW42111.1 AJWK01006903 GGFJ01002559 MBW51700.1 KQ982409 KYQ56647.1 AXCM01000884 GECZ01008841 JAS60928.1 ADMH02001371 ETN62749.1 AAAB01008859 EAA07937.5 APCN01000612 GDHF01032141 GDHF01015087 GDHF01009790 GDHF01003337 JAI20173.1 JAI37227.1 JAI42524.1 JAI48977.1 GAKP01011728 GAKP01011727 GAKP01011726 GAKP01011725 JAC47227.1 GFDF01008447 JAV05637.1 CH477356 EAT42772.1 CVRI01000020 CRK90547.1 JRES01001143 KNC25160.1 GFDL01001655 JAV33390.1 JXJN01021534 DS235758 EEB16461.1 GGFK01009152 MBW42473.1 CH933811 EDW06344.1 CH954180 EDV47688.1 CM002911 KMY91726.1 ABLF02031355 X62679 AE014298 BT012671 BT021384 AY102664 CH902630 KPU77416.1 CH891579 KRK05040.1 CH480924 EDW43670.1 CH964239 EDW82265.1 CP012528 ALC49674.1 ADTU01015668 ADTU01015669 ADTU01015670 ADTU01015671 ADTU01015672 ADTU01015673 ADTU01015674 ADTU01015675 ADTU01015676 GGMS01014519 MBY83722.1 CH379064 EDY72500.2 GGFJ01003020 MBW52161.1 GEHC01001212 JAV46433.1 AY347747 AAR01873.1 OUUW01000025 SPP89644.1 AF097830 CH940653 AAC99436.1 KRF80528.1 CH916370 EDW00078.1 CH479209 EDW32721.1 DS231864 EDS40213.1 LNIX01000002 OXA60580.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000076502
+ More
UP000053105 UP000005203 UP000053825 UP000000311 UP000027135 UP000215335 UP000053097 UP000279307 UP000078540 UP000078492 UP000007755 UP000008237 UP000078542 UP000015103 UP000192223 UP000242457 UP000036403 UP000078541 UP000007266 UP000019118 UP000030742 UP000076408 UP000075881 UP000075885 UP000075880 UP000075900 UP000075886 UP000092462 UP000030765 UP000069272 UP000092461 UP000075809 UP000075883 UP000000673 UP000075903 UP000076407 UP000007062 UP000075840 UP000075902 UP000075884 UP000095301 UP000008820 UP000091820 UP000183832 UP000092445 UP000037069 UP000092443 UP000095300 UP000092460 UP000009046 UP000009192 UP000008711 UP000007819 UP000000803 UP000007801 UP000002282 UP000001292 UP000192221 UP000007798 UP000092553 UP000005205 UP000001819 UP000268350 UP000008792 UP000001070 UP000008744 UP000075920 UP000002320 UP000198287
UP000053105 UP000005203 UP000053825 UP000000311 UP000027135 UP000215335 UP000053097 UP000279307 UP000078540 UP000078492 UP000007755 UP000008237 UP000078542 UP000015103 UP000192223 UP000242457 UP000036403 UP000078541 UP000007266 UP000019118 UP000030742 UP000076408 UP000075881 UP000075885 UP000075880 UP000075900 UP000075886 UP000092462 UP000030765 UP000069272 UP000092461 UP000075809 UP000075883 UP000000673 UP000075903 UP000076407 UP000007062 UP000075840 UP000075902 UP000075884 UP000095301 UP000008820 UP000091820 UP000183832 UP000092445 UP000037069 UP000092443 UP000095300 UP000092460 UP000009046 UP000009192 UP000008711 UP000007819 UP000000803 UP000007801 UP000002282 UP000001292 UP000192221 UP000007798 UP000092553 UP000005205 UP000001819 UP000268350 UP000008792 UP000001070 UP000008744 UP000075920 UP000002320 UP000198287
Interpro
SUPFAM
SSF48452
SSF48452
ProteinModelPortal
H9JNM9
A0A2A4IZY6
A0A194PSH1
A0A2W1BC53
A0A212FJS9
A0A2H1WFW9
+ More
A0A194RMW6 A0A154PG94 A0A0C9S0B0 A0A0M8ZTK3 A0A310S893 A0A088AKK0 A0A0L7R9J2 E2AWC7 A0A067R195 A0A232ENU7 A0A026X3Y1 A0A3L8DG90 A0A195AZG7 A0A195EE00 F4X1W4 E2BXK4 A0A195D4K0 A0A1B6CGC0 T1HT88 A0A1W4X9X1 A0A023F598 A0A2A3EH94 A0A0J7KXU5 A0A1B6KX04 A0A195FYA3 D2A3P0 A0A0A9YM07 N6SVB2 A0A1B6K0U8 A0A1B6ENK3 A0A182Y4B2 A0A182JTF5 A0A182P5T0 A0A182IRV9 A0A182RNQ7 A0A182QHX7 A0A1B0DCA6 A0A1B6J9Z5 A0A084VRC4 A0A2M4AMV5 A0A182F7E1 A0A1B0CCL9 A0A2M4BF68 A0A151X8E4 A0A182MUI8 A0A1B6GER8 W5JIY1 A0A182VE99 A0A182WRJ2 Q7QDA0 A0A182HU85 A0A182TDN5 A0A182N635 A0A1S4FBE7 A0A0K8VUG6 A0A034VYT8 A0A1L8DGX8 A0A1I8M8R5 Q0IFE2 A0A1A9W3Z9 A0A1J1HRE8 A0A1B0AC18 A0A0L0BYQ7 A0A1A9XE12 A0A1I8M8S0 A0A1I8PF42 A0A1Q3G0R8 A0A1B0BVY6 E0VSV5 A0A2M4ANX4 B4L585 B3NYA3 A0A0J9R6P7 J9JQD1 P25991-4 A0A0P8XS09 A0A0R1E736 B4IM34 A0A1W4WDD0 B4NEU6 A0A0M5J1D0 A0A158NHM7 A0A2S2R309 B5DLV3 A0A2M4BGT1 A0A1W7R5N0 Q6V8Y2 A0A3B0K687 O96641 B4JKH5 B4H4H0 A0A182W069 B0W9I8 A0A226ETM0
A0A194RMW6 A0A154PG94 A0A0C9S0B0 A0A0M8ZTK3 A0A310S893 A0A088AKK0 A0A0L7R9J2 E2AWC7 A0A067R195 A0A232ENU7 A0A026X3Y1 A0A3L8DG90 A0A195AZG7 A0A195EE00 F4X1W4 E2BXK4 A0A195D4K0 A0A1B6CGC0 T1HT88 A0A1W4X9X1 A0A023F598 A0A2A3EH94 A0A0J7KXU5 A0A1B6KX04 A0A195FYA3 D2A3P0 A0A0A9YM07 N6SVB2 A0A1B6K0U8 A0A1B6ENK3 A0A182Y4B2 A0A182JTF5 A0A182P5T0 A0A182IRV9 A0A182RNQ7 A0A182QHX7 A0A1B0DCA6 A0A1B6J9Z5 A0A084VRC4 A0A2M4AMV5 A0A182F7E1 A0A1B0CCL9 A0A2M4BF68 A0A151X8E4 A0A182MUI8 A0A1B6GER8 W5JIY1 A0A182VE99 A0A182WRJ2 Q7QDA0 A0A182HU85 A0A182TDN5 A0A182N635 A0A1S4FBE7 A0A0K8VUG6 A0A034VYT8 A0A1L8DGX8 A0A1I8M8R5 Q0IFE2 A0A1A9W3Z9 A0A1J1HRE8 A0A1B0AC18 A0A0L0BYQ7 A0A1A9XE12 A0A1I8M8S0 A0A1I8PF42 A0A1Q3G0R8 A0A1B0BVY6 E0VSV5 A0A2M4ANX4 B4L585 B3NYA3 A0A0J9R6P7 J9JQD1 P25991-4 A0A0P8XS09 A0A0R1E736 B4IM34 A0A1W4WDD0 B4NEU6 A0A0M5J1D0 A0A158NHM7 A0A2S2R309 B5DLV3 A0A2M4BGT1 A0A1W7R5N0 Q6V8Y2 A0A3B0K687 O96641 B4JKH5 B4H4H0 A0A182W069 B0W9I8 A0A226ETM0
PDB
2OOE
E-value=0,
Score=1925
Ontologies
GO
Topology
Subcellular location
Nucleus
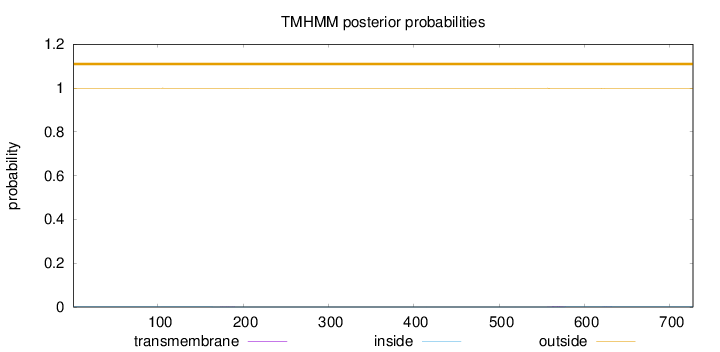
Length:
727
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03521
Exp number, first 60 AAs:
0.00034
Total prob of N-in:
0.00071
outside
1 - 727
Population Genetic Test Statistics
Pi
17.086106
Theta
17.159136
Tajima's D
-1.469072
CLR
111.892458
CSRT
0.0620968951552422
Interpretation
Uncertain
