Gene
KWMTBOMO14184 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011132
Annotation
U6_snRNA-associated_Sm-like_protein_LSm2_[Operophtera_brumata]
Full name
U6 snRNA-associated Sm-like protein LSm2
Location in the cell
Nuclear Reliability : 1.832
Sequence
CDS
ATGTTGTTTTACTCGTTTTTCAAATCGTTAGTCGGTAAGGATGTAGTCGTCGAGCTGAAAAATGACCTCAGCATATGTGGGACGTTGCATTCTGTGGATCAATATCTCAATATAAAGCTTTCGGATATAAGCGTGATTGACTCTGAAAAGTATCCTCATATGTTATCAGTGAAAAATTGTTTTATAAGAGGCTCCGTGGTTCGATACGTACAACTGCCGGCTGATGAGGTAGACACCCAGCTTCTCCAGGATGCCGCTAGAAAGGAGGCGACCGTTTCAACAAGATAG
Protein
MLFYSFFKSLVGKDVVVELKNDLSICGTLHSVDQYLNIKLSDISVIDSEKYPHMLSVKNCFIRGSVVRYVQLPADEVDTQLLQDAARKEATVSTR
Summary
Description
Binds specifically to the 3'-terminal U-tract of U6 snRNA.
Similarity
Belongs to the snRNP Sm proteins family.
Uniprot
A0A0L7L5U9
A0A2W1BCZ8
A0A2H1WFD9
S4PPF6
A0A194PZ02
D6WBR3
+ More
A0A1A9WD03 A0A1W4XH99 B4L066 B3M7T6 A0A0L0BWN0 A0A2R5LG58 A0A1B0G3P4 A0A1A9YKR5 A0A1A9URT5 A0A1B0C0C6 A0A1Z5LFB2 N6TPC3 A0A1W4UGX3 B4LFF8 Q9VTW6 B4PGC1 B4N3J0 B4HFL6 B3NH93 B4QR91 Q5RKP8 W8C048 A0A1Q3FUS5 A0A182YNR3 A0A182Q0W0 A0A182V4V9 A0A182N7Z4 Q17LD2 A0A182LBE4 A0A182XTH7 Q7PQ03 A0A182HP54 A0A023ECH8 A0A0K8TRI8 A0A1B0D5H2 A0A1B0GI52 A0A0P4VYA5 A0A293MHL1 A0A3Q7RKR1 A0A3B0KQC5 Q2M0N7 B4GRD5 B7QF41 A0A0K8RDN5 A0A2S2N493 U5ELI6 A0A182WAC1 A0A2M4AZC3 A0A2M4C4Q8 W5J9N6 A0A182FH97 A0A3Q3S0G5 A0A067QFW5 A0A3P8UQ36 A0A3B4WG76 A0A3B3XPE5 A0A3B4GWC9 A0A3Q1GAB9 A0A3B5APL1 A0A3Q3DZT1 A0A2I4CCT9 W5MD35 A0A3B3RYH1 G3NI70 A0A3Q1JNJ5 A0A087X6Y2 A0A3B3BAA6 A0A3B4VH84 A0A3Q3ASV6 A0A3Q3K346 A0A3B3VTF3 I3KT75 A0A0E9X3P2 A0A3B1IFD5 A0A3B3IAS1 A0A3B4DCP3 A0A3P8R3F5 H3B5C4 A0A1A8R8A5 A0A1A8QJF2 A0A1A8IAL9 A0A1A7YRD4 A0A0P7Z647 E3TF96 A0A1A8E4J3 A0A1A8FP03 A0A1A8V8Q2 B4J336 C1C4Z7 Q52KE0 Q5BL46 A0A2U9C6S1 V9LKC1 V9ILZ7
A0A1A9WD03 A0A1W4XH99 B4L066 B3M7T6 A0A0L0BWN0 A0A2R5LG58 A0A1B0G3P4 A0A1A9YKR5 A0A1A9URT5 A0A1B0C0C6 A0A1Z5LFB2 N6TPC3 A0A1W4UGX3 B4LFF8 Q9VTW6 B4PGC1 B4N3J0 B4HFL6 B3NH93 B4QR91 Q5RKP8 W8C048 A0A1Q3FUS5 A0A182YNR3 A0A182Q0W0 A0A182V4V9 A0A182N7Z4 Q17LD2 A0A182LBE4 A0A182XTH7 Q7PQ03 A0A182HP54 A0A023ECH8 A0A0K8TRI8 A0A1B0D5H2 A0A1B0GI52 A0A0P4VYA5 A0A293MHL1 A0A3Q7RKR1 A0A3B0KQC5 Q2M0N7 B4GRD5 B7QF41 A0A0K8RDN5 A0A2S2N493 U5ELI6 A0A182WAC1 A0A2M4AZC3 A0A2M4C4Q8 W5J9N6 A0A182FH97 A0A3Q3S0G5 A0A067QFW5 A0A3P8UQ36 A0A3B4WG76 A0A3B3XPE5 A0A3B4GWC9 A0A3Q1GAB9 A0A3B5APL1 A0A3Q3DZT1 A0A2I4CCT9 W5MD35 A0A3B3RYH1 G3NI70 A0A3Q1JNJ5 A0A087X6Y2 A0A3B3BAA6 A0A3B4VH84 A0A3Q3ASV6 A0A3Q3K346 A0A3B3VTF3 I3KT75 A0A0E9X3P2 A0A3B1IFD5 A0A3B3IAS1 A0A3B4DCP3 A0A3P8R3F5 H3B5C4 A0A1A8R8A5 A0A1A8QJF2 A0A1A8IAL9 A0A1A7YRD4 A0A0P7Z647 E3TF96 A0A1A8E4J3 A0A1A8FP03 A0A1A8V8Q2 B4J336 C1C4Z7 Q52KE0 Q5BL46 A0A2U9C6S1 V9LKC1 V9ILZ7
Pubmed
26227816
28756777
23622113
26354079
18362917
19820115
+ More
17994087 26108605 28528879 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 23594743 24495485 25244985 17510324 20966253 12364791 14747013 17210077 24945155 26483478 26369729 27129103 15632085 20920257 23761445 24845553 24487278 29240929 29451363 25186727 25613341 25329095 17554307 9215903 20634964 27762356 20431018 24402279
17994087 26108605 28528879 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 23594743 24495485 25244985 17510324 20966253 12364791 14747013 17210077 24945155 26483478 26369729 27129103 15632085 20920257 23761445 24845553 24487278 29240929 29451363 25186727 25613341 25329095 17554307 9215903 20634964 27762356 20431018 24402279
EMBL
JTDY01002804
KOB70686.1
KZ150147
PZC72868.1
ODYU01008301
SOQ51771.1
+ More
GAIX01000277 JAA92283.1 KQ459593 KPI96385.1 KQ971314 EEZ97873.1 CH933809 EDW18012.1 CH902618 EDV38809.2 JRES01001238 KNC24416.1 GGLE01004375 MBY08501.1 CCAG010015898 JXJN01023513 GFJQ02001276 JAW05694.1 APGK01026590 KB740635 KB631996 KI210453 ENN79873.1 ERL87789.1 ERL96271.1 CH940647 EDW69256.1 AE014296 AAF49929.1 CM000159 EDW94285.1 CH964095 EDW79195.2 CH480815 EDW41247.1 CH954178 EDV51550.2 CM000363 CM002912 EDX10221.1 KMY99215.1 FP101882 BC085441 BC152150 BC164435 AAH85441.1 AAI52151.1 AAI64435.1 GAMC01001528 JAC05028.1 GFDL01003740 JAV31305.1 AXCN02000909 CH477216 EAT47530.1 AAAB01008900 EAA09471.3 APCN01002921 JXUM01123609 GAPW01006546 KQ566736 JAC07052.1 KXJ69874.1 GDAI01000850 JAI16753.1 AJVK01025246 AJWK01011307 GDKW01000992 JAI55603.1 GFWV01015177 MAA39906.1 OUUW01000012 SPP87411.1 CH379069 EAL30893.1 CH479188 EDW40320.1 ABJB010168149 ABJB010353024 DS923918 EEC17463.1 GADI01004632 GEFM01003833 JAA69176.1 JAP71963.1 GGMT01000287 MBY11973.1 GANO01001390 JAB58481.1 GGFK01012825 MBW46146.1 GGFJ01011094 MBW60235.1 ADMH02002078 ETN59459.1 KK869568 KDR06681.1 AHAT01015189 AYCK01014133 AERX01011533 GBXM01011245 JAH97332.1 AFYH01090908 AFYH01090909 HAEH01015127 HAEI01008199 SBS01922.1 HAEF01008255 HAEG01012783 SBR93234.1 HAED01007945 HAEE01012874 SBQ94157.1 HADW01004822 HADX01010529 SBP32761.1 JARO02001213 KPP76144.1 GU589030 ADO28982.1 HADZ01010773 HAEA01011529 SBQ40009.1 HAEB01014588 HAEC01005412 SBQ61115.1 HADY01016841 HAEJ01015500 SBS55957.1 CH916366 EDV96107.1 BT081926 ACO52057.1 BC094397 CM004481 AAH94397.1 OCT66214.1 AAMC01092845 BC090606 CR761142 AAH90606.1 CAJ81423.1 CP026254 AWP11426.1 JW881425 AFP13942.1 JR050874 AEY61374.1
GAIX01000277 JAA92283.1 KQ459593 KPI96385.1 KQ971314 EEZ97873.1 CH933809 EDW18012.1 CH902618 EDV38809.2 JRES01001238 KNC24416.1 GGLE01004375 MBY08501.1 CCAG010015898 JXJN01023513 GFJQ02001276 JAW05694.1 APGK01026590 KB740635 KB631996 KI210453 ENN79873.1 ERL87789.1 ERL96271.1 CH940647 EDW69256.1 AE014296 AAF49929.1 CM000159 EDW94285.1 CH964095 EDW79195.2 CH480815 EDW41247.1 CH954178 EDV51550.2 CM000363 CM002912 EDX10221.1 KMY99215.1 FP101882 BC085441 BC152150 BC164435 AAH85441.1 AAI52151.1 AAI64435.1 GAMC01001528 JAC05028.1 GFDL01003740 JAV31305.1 AXCN02000909 CH477216 EAT47530.1 AAAB01008900 EAA09471.3 APCN01002921 JXUM01123609 GAPW01006546 KQ566736 JAC07052.1 KXJ69874.1 GDAI01000850 JAI16753.1 AJVK01025246 AJWK01011307 GDKW01000992 JAI55603.1 GFWV01015177 MAA39906.1 OUUW01000012 SPP87411.1 CH379069 EAL30893.1 CH479188 EDW40320.1 ABJB010168149 ABJB010353024 DS923918 EEC17463.1 GADI01004632 GEFM01003833 JAA69176.1 JAP71963.1 GGMT01000287 MBY11973.1 GANO01001390 JAB58481.1 GGFK01012825 MBW46146.1 GGFJ01011094 MBW60235.1 ADMH02002078 ETN59459.1 KK869568 KDR06681.1 AHAT01015189 AYCK01014133 AERX01011533 GBXM01011245 JAH97332.1 AFYH01090908 AFYH01090909 HAEH01015127 HAEI01008199 SBS01922.1 HAEF01008255 HAEG01012783 SBR93234.1 HAED01007945 HAEE01012874 SBQ94157.1 HADW01004822 HADX01010529 SBP32761.1 JARO02001213 KPP76144.1 GU589030 ADO28982.1 HADZ01010773 HAEA01011529 SBQ40009.1 HAEB01014588 HAEC01005412 SBQ61115.1 HADY01016841 HAEJ01015500 SBS55957.1 CH916366 EDV96107.1 BT081926 ACO52057.1 BC094397 CM004481 AAH94397.1 OCT66214.1 AAMC01092845 BC090606 CR761142 AAH90606.1 CAJ81423.1 CP026254 AWP11426.1 JW881425 AFP13942.1 JR050874 AEY61374.1
Proteomes
UP000037510
UP000053268
UP000007266
UP000091820
UP000192223
UP000009192
+ More
UP000007801 UP000037069 UP000092444 UP000092443 UP000078200 UP000092460 UP000019118 UP000030742 UP000192221 UP000008792 UP000000803 UP000002282 UP000007798 UP000001292 UP000008711 UP000000304 UP000000437 UP000076408 UP000075886 UP000075903 UP000075884 UP000008820 UP000075882 UP000076407 UP000007062 UP000075840 UP000069940 UP000249989 UP000092462 UP000092461 UP000286640 UP000268350 UP000001819 UP000008744 UP000001555 UP000075920 UP000000673 UP000069272 UP000261640 UP000027135 UP000265120 UP000261360 UP000261480 UP000261460 UP000257200 UP000261400 UP000264820 UP000192220 UP000018468 UP000261540 UP000007635 UP000265040 UP000028760 UP000261560 UP000261420 UP000264800 UP000261600 UP000261500 UP000005207 UP000018467 UP000001038 UP000261440 UP000265100 UP000008672 UP000034805 UP000192224 UP000221080 UP000001070 UP000186698 UP000008143 UP000246464
UP000007801 UP000037069 UP000092444 UP000092443 UP000078200 UP000092460 UP000019118 UP000030742 UP000192221 UP000008792 UP000000803 UP000002282 UP000007798 UP000001292 UP000008711 UP000000304 UP000000437 UP000076408 UP000075886 UP000075903 UP000075884 UP000008820 UP000075882 UP000076407 UP000007062 UP000075840 UP000069940 UP000249989 UP000092462 UP000092461 UP000286640 UP000268350 UP000001819 UP000008744 UP000001555 UP000075920 UP000000673 UP000069272 UP000261640 UP000027135 UP000265120 UP000261360 UP000261480 UP000261460 UP000257200 UP000261400 UP000264820 UP000192220 UP000018468 UP000261540 UP000007635 UP000265040 UP000028760 UP000261560 UP000261420 UP000264800 UP000261600 UP000261500 UP000005207 UP000018467 UP000001038 UP000261440 UP000265100 UP000008672 UP000034805 UP000192224 UP000221080 UP000001070 UP000186698 UP000008143 UP000246464
PRIDE
Pfam
PF01423 LSM
SUPFAM
SSF50182
SSF50182
CDD
ProteinModelPortal
A0A0L7L5U9
A0A2W1BCZ8
A0A2H1WFD9
S4PPF6
A0A194PZ02
D6WBR3
+ More
A0A1A9WD03 A0A1W4XH99 B4L066 B3M7T6 A0A0L0BWN0 A0A2R5LG58 A0A1B0G3P4 A0A1A9YKR5 A0A1A9URT5 A0A1B0C0C6 A0A1Z5LFB2 N6TPC3 A0A1W4UGX3 B4LFF8 Q9VTW6 B4PGC1 B4N3J0 B4HFL6 B3NH93 B4QR91 Q5RKP8 W8C048 A0A1Q3FUS5 A0A182YNR3 A0A182Q0W0 A0A182V4V9 A0A182N7Z4 Q17LD2 A0A182LBE4 A0A182XTH7 Q7PQ03 A0A182HP54 A0A023ECH8 A0A0K8TRI8 A0A1B0D5H2 A0A1B0GI52 A0A0P4VYA5 A0A293MHL1 A0A3Q7RKR1 A0A3B0KQC5 Q2M0N7 B4GRD5 B7QF41 A0A0K8RDN5 A0A2S2N493 U5ELI6 A0A182WAC1 A0A2M4AZC3 A0A2M4C4Q8 W5J9N6 A0A182FH97 A0A3Q3S0G5 A0A067QFW5 A0A3P8UQ36 A0A3B4WG76 A0A3B3XPE5 A0A3B4GWC9 A0A3Q1GAB9 A0A3B5APL1 A0A3Q3DZT1 A0A2I4CCT9 W5MD35 A0A3B3RYH1 G3NI70 A0A3Q1JNJ5 A0A087X6Y2 A0A3B3BAA6 A0A3B4VH84 A0A3Q3ASV6 A0A3Q3K346 A0A3B3VTF3 I3KT75 A0A0E9X3P2 A0A3B1IFD5 A0A3B3IAS1 A0A3B4DCP3 A0A3P8R3F5 H3B5C4 A0A1A8R8A5 A0A1A8QJF2 A0A1A8IAL9 A0A1A7YRD4 A0A0P7Z647 E3TF96 A0A1A8E4J3 A0A1A8FP03 A0A1A8V8Q2 B4J336 C1C4Z7 Q52KE0 Q5BL46 A0A2U9C6S1 V9LKC1 V9ILZ7
A0A1A9WD03 A0A1W4XH99 B4L066 B3M7T6 A0A0L0BWN0 A0A2R5LG58 A0A1B0G3P4 A0A1A9YKR5 A0A1A9URT5 A0A1B0C0C6 A0A1Z5LFB2 N6TPC3 A0A1W4UGX3 B4LFF8 Q9VTW6 B4PGC1 B4N3J0 B4HFL6 B3NH93 B4QR91 Q5RKP8 W8C048 A0A1Q3FUS5 A0A182YNR3 A0A182Q0W0 A0A182V4V9 A0A182N7Z4 Q17LD2 A0A182LBE4 A0A182XTH7 Q7PQ03 A0A182HP54 A0A023ECH8 A0A0K8TRI8 A0A1B0D5H2 A0A1B0GI52 A0A0P4VYA5 A0A293MHL1 A0A3Q7RKR1 A0A3B0KQC5 Q2M0N7 B4GRD5 B7QF41 A0A0K8RDN5 A0A2S2N493 U5ELI6 A0A182WAC1 A0A2M4AZC3 A0A2M4C4Q8 W5J9N6 A0A182FH97 A0A3Q3S0G5 A0A067QFW5 A0A3P8UQ36 A0A3B4WG76 A0A3B3XPE5 A0A3B4GWC9 A0A3Q1GAB9 A0A3B5APL1 A0A3Q3DZT1 A0A2I4CCT9 W5MD35 A0A3B3RYH1 G3NI70 A0A3Q1JNJ5 A0A087X6Y2 A0A3B3BAA6 A0A3B4VH84 A0A3Q3ASV6 A0A3Q3K346 A0A3B3VTF3 I3KT75 A0A0E9X3P2 A0A3B1IFD5 A0A3B3IAS1 A0A3B4DCP3 A0A3P8R3F5 H3B5C4 A0A1A8R8A5 A0A1A8QJF2 A0A1A8IAL9 A0A1A7YRD4 A0A0P7Z647 E3TF96 A0A1A8E4J3 A0A1A8FP03 A0A1A8V8Q2 B4J336 C1C4Z7 Q52KE0 Q5BL46 A0A2U9C6S1 V9LKC1 V9ILZ7
PDB
6QX9
E-value=4.35831e-42,
Score=424
Ontologies
PATHWAY
GO
PANTHER
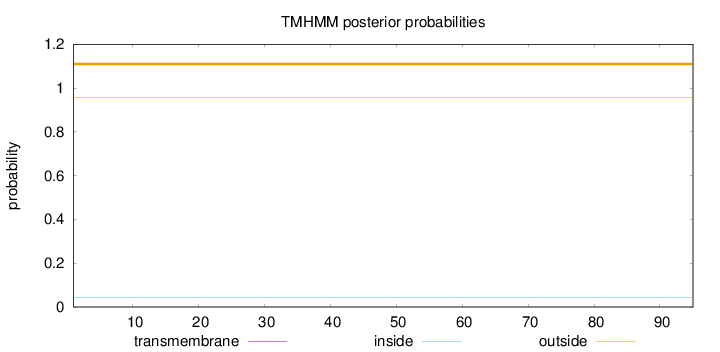
Topology
Length:
95
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.04420
outside
1 - 95
Population Genetic Test Statistics
Pi
11.113232
Theta
11.435017
Tajima's D
-0.083331
CLR
0
CSRT
0.357582120893955
Interpretation
Uncertain
