Pre Gene Modal
BGIBMGA011132
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_ankyrin_repeat_domain-containing_protein_50-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.096 Nuclear Reliability : 1.347
Sequence
CDS
ATGAATGCTGCGAATGAATTGATATTTCTAGTTCTGGACGGTGTCGCAGACGGCTTCATAGCTCTTCGTGAGATTAGGGAAATACCCAGCACTCTGAACGGATTGTATCTGTGGCTGGCGCAGAGACTGTTCCATGGGAGACGATTCGCTAAGGTTCGTCTCCTGCTGGACGTGCTGCTGGCGGCTCGGTGCGGGGTCACGGAGGAGATGCTCTACAAATGTCTGCTCACGAAGGAATACAGTGTAACACGAGAAGATTTCAATAGAAGAATTCACTTGCTCAGGAGAATCCTGGTGATTGAGAAGAGCACGGGCTACCTGTCGATATTCCACCACTCGTTCTCTTGCTGGCTGCTGGACGTCAAGCACTGCACGCGGCGGTACCTGTGTTGCCTGGCGGATGGGCACGCCGCGCTCGCCATGTACTACACACTCATAGCGAAACGCCTGTCGGCGCTGGAGATCCACAACTACGTGTACCACATGACGCGGCTGGAGCATCATCTGTCGTCGCTGAATAAGAGCAGAGGTGATCAGGATGGGCCGGTCGATCTGCACACTTTGGTCTTGTTGTGGGTGTTGGATTCTTCGTGCGACGTCGAGTCGGCCCTCAAGAACGAGGTCCGGAGACTGGACATGTCGCAGAAAACTGAAGCGAGCCACGTCGAAGAGTCGAGCGGGAGGGAGGAGAACGCGCAAACTACAGATTCCGCCTCAATTGAGTCAATAATGCCGGAGCTGATCAGCGGAGCCCCAGTCGCTTGGTGCAAAGACAAGCGGGTCGCGAGGGCGCTCTACGAACTAGCTAAAGGTCCGGCCCTGCTCGACCCCAGCGACGTCGGAGCCGGAGAGGTGATAATAGAGCAGTCGAACGAGGATGAGGTCCCACAGGAATCTTGTGAGGAACGAGAGGATCCCATAGTCCTGGATCCGAATATCATCCACGAGCTGGTCGCTAGGGCGTCACCCGGAGCTGGTGGGCGCGGGGGCGGTGGGGAGGCGGGGCTGCAGGCGGCGGCGGCGCGCGGGCGGGCGGGCTGCGCGCTGGCCCTGCTGCGTGCGGGGGCGGCGCCCGACGTGCCTGACGCTGACGGCTGGAGCCCGCTCCGAGCCGCCGCCTGGGCTGGACACACCGAGGTTGTGGACGTGCTGCTGGAGTACGGATGTGACGTCGATTGTGTGGACGCGGACAACCGGACCGCGCTGAGGGCGGCGGCGTGGTCCGGCCACGAGGCGGTGGTCTCCCGGCTGCTGGCCGCCGGCGCAGCCCCGGACCGCGCGGACGCGGAGGGCCGCACGGCGCTGATAGCCGCCGCGTACATGGGGCACGCCGAGATCGTGAGGGCGCTGCTGGACGCCGGGGCCCGCGTGGACCACGCCGACGAGGACGGTCGCACGGCGCTGGCGGTGGCGTCGCTGTGCGCGGGCGGCGCGGCGTGCGCGGCGCTGCTGCTGGAGCGCGGCGCTGACGTCACGGCGCACGACCGCGACCACGCCACGCCGCTGCTCGTCGCCGCCTTCGAGGGTCACACCGAGATCTGCGAGTTGCTGCTGGAGGCGGAGGCGGACATCGAGGCGTGCGACGGCGCCGGGCGCACCGCGCTGTGGGCGGCGGCGTCGGCCGGGCACGCCGACACCGTGCGCCTGCTGCTGTTCTGGGGCGCCTGCGTGGACGGCATGGACGCCGAGGGCCGCACCGTGCTGTGCACCGCCGCCGCGCAGGGCAACGTGGAAGTGGTGCGGCAACTCCTCGACCGCGGCCTGGACGAGCACCACCGTGACAACTCCGGGTGGACGCCCCTGCATTACGCCGCTTTTGAAGGTCACATCGAAGTGTGCGAAGCGTTACTAGAGGCAGGCGCTAAGGTGGACGAGGCCGACAACGACGGTAAGGGGGCGCTGATGCTGGCCGCCCAGGAGGGACACACCAGGCTGGTGGAGATCCTCGTGGACGACTGGGGGGCGCCCGTCGACCAGAGAGCTCACGACGGGAAGACCGCTTTAAGGCTGGCTGCTCTGGAGGGCCAGTTCTCGACGGTGTCGGCGCTGCACGCGCGCGGCGCGGACGCGGACGCCGTGGACGCGGACCGGCGCAGCACCCTGTACGTGCTGGCCCTCGACAACCGCCTCGCCATGGCGCGCTTCCTGCTGACCTCCTGCGGCGCCAGCGTGCACACGGCCGATGCTGAGGTTCGGACTCCCTTGCACGTATCAGCGTGGCAGGGTCACACCGACATGGTGAATTTGCTGATCACAGTAGGAGGGGCGCGCGCGGACGGGCGGGACGCGGCGCGGCGCACGGCGCTGCACGCGGCGGCGTGGCGCGGGCGGCGGGCGGCGCTGCACGCGCTGCTGCGGCACGGCGCCGACCCCTCCGCACTGTGCTCGCAGGGAGCCACGCCGCTAGGTATAGCGGCCCAGGAAGGTCACGAGGAGTGCGTCCTGTATTTGCTGCAGTATGGCGCTGATCCCCAGCAGACCGACCACTGTGGTCGGACGCCTGCGAAAGTCGCCTGGCGGGCGGGCCACGCGAACATATGTCGCTTGTTGGAACGGTGGCCGACGGTCAGCGAACAGCTACCACCGACGGCGCCGCCGCTGGCCGCCAGAACGCCCAGTGCTGGCTCACCGGAGTTCAAACGCCGCAGCGTCCATAGTTCGAACTCGACCAAATCCTCGTCGAACCTGACCGGGGGCTCGCACCCGTCGCACGAGCCCGAGCCGCCCGCGGAGCCCCACGACAAGGGCCGCGCGAGTCTGTCGTTCGCGCAGCAAGTGGCGCGCTGCGGGCGGCCGCGCCGCGGGCGCGAGCGGGACAGCACGATACCGGAGCACGCGGCTGCCGAGGACGACGCCAAGCTGAGGAGTTACATAGCGCACGAGCGAGACGCCGAGCTCCGCGGTTACGTGTGCGAGCGGGACAGGCGGCGCGACGCTCGCCACGGGTGCGAGCGGGACAGCACGAGCCCGCTGTACGCGTCGCCGCCGCGGAGTCCCACCAGCGACATCTGCAGCCCTACACAACCTACTGTAGCTTTTGGATCCCAGCCGCCGAGCCTGTCGACCGTCCAGCCCGGGCTCGCGGACCCTCACTTCAACAGAGACACGCACATGAGGATCATACTGGGCAGGGACAGCAAACCGGCTCACGATAAAAACGACAGTGCATTGTGGACGACGAAAGGAAGGCTAGCCTGCAACAAGTTATTCGGGAGTATCGAGAGAACGAGAGGCAGGATATTAAAGAAAACCAAGAACAAAAGAAATGGAATCGTCACGAATCCAGCGATGAGATTGGTGGCGAACGTCAGGAATGGACTCGATAACGCGGCGGCCAACATCCGGAGGACCGGCGTCGCCCTGGCGGCCAGCACTAGCTCCAACAATCCGGCAGTCAAGTCCAACGCGTTCCAGTGGAGGAAGGAGACGCCGTTGTAA
Protein
MNAANELIFLVLDGVADGFIALREIREIPSTLNGLYLWLAQRLFHGRRFAKVRLLLDVLLAARCGVTEEMLYKCLLTKEYSVTREDFNRRIHLLRRILVIEKSTGYLSIFHHSFSCWLLDVKHCTRRYLCCLADGHAALAMYYTLIAKRLSALEIHNYVYHMTRLEHHLSSLNKSRGDQDGPVDLHTLVLLWVLDSSCDVESALKNEVRRLDMSQKTEASHVEESSGREENAQTTDSASIESIMPELISGAPVAWCKDKRVARALYELAKGPALLDPSDVGAGEVIIEQSNEDEVPQESCEEREDPIVLDPNIIHELVARASPGAGGRGGGGEAGLQAAAARGRAGCALALLRAGAAPDVPDADGWSPLRAAAWAGHTEVVDVLLEYGCDVDCVDADNRTALRAAAWSGHEAVVSRLLAAGAAPDRADAEGRTALIAAAYMGHAEIVRALLDAGARVDHADEDGRTALAVASLCAGGAACAALLLERGADVTAHDRDHATPLLVAAFEGHTEICELLLEAEADIEACDGAGRTALWAAASAGHADTVRLLLFWGACVDGMDAEGRTVLCTAAAQGNVEVVRQLLDRGLDEHHRDNSGWTPLHYAAFEGHIEVCEALLEAGAKVDEADNDGKGALMLAAQEGHTRLVEILVDDWGAPVDQRAHDGKTALRLAALEGQFSTVSALHARGADADAVDADRRSTLYVLALDNRLAMARFLLTSCGASVHTADAEVRTPLHVSAWQGHTDMVNLLITVGGARADGRDAARRTALHAAAWRGRRAALHALLRHGADPSALCSQGATPLGIAAQEGHEECVLYLLQYGADPQQTDHCGRTPAKVAWRAGHANICRLLERWPTVSEQLPPTAPPLAARTPSAGSPEFKRRSVHSSNSTKSSSNLTGGSHPSHEPEPPAEPHDKGRASLSFAQQVARCGRPRRGRERDSTIPEHAAAEDDAKLRSYIAHERDAELRGYVCERDRRRDARHGCERDSTSPLYASPPRSPTSDICSPTQPTVAFGSQPPSLSTVQPGLADPHFNRDTHMRIILGRDSKPAHDKNDSALWTTKGRLACNKLFGSIERTRGRILKKTKNKRNGIVTNPAMRLVANVRNGLDNAAANIRRTGVALAASTSSNNPAVKSNAFQWRKETPL
Summary
Uniprot
Proteomes
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
PDB
6MOL
E-value=2.61576e-46,
Score=472
Ontologies
GO
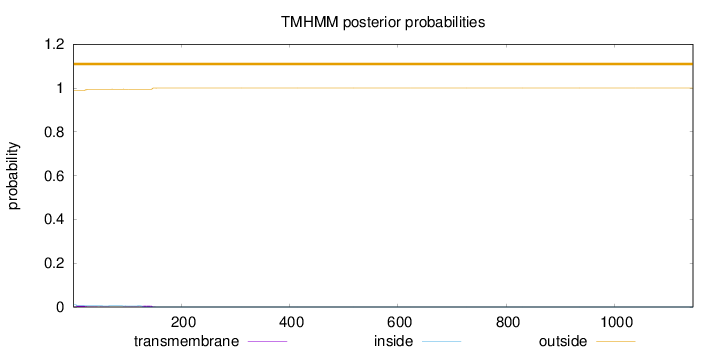
Topology
Length:
1145
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.232090000000001
Exp number, first 60 AAs:
0.10143
Total prob of N-in:
0.01052
outside
1 - 1145
Population Genetic Test Statistics
Pi
23.079854
Theta
18.862204
Tajima's D
-1.445396
CLR
108.220596
CSRT
0.0652467376631168
Interpretation
Uncertain
