Gene
KWMTBOMO14178
Pre Gene Modal
BGIBMGA011100
Annotation
PREDICTED:_leucine-rich_repeat_neuronal_protein_1-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.238 PlasmaMembrane Reliability : 1.837
Sequence
CDS
ATGTTTTACTGTAAATATATGTTTATTATTTTATTCTTATTAACGAGAACACAAAAACAAATTAGTCAAGCAGCAGAATTACAAGCTATTGAGCAGACAGGAACGGTTACAGAAATCATTCCATCAGTTACAGTCAGTGCATTAGCCGATTTCGAAAGCAATGGAGAAACTACCACAGATGTTGCGGTGTTAAAGCAAGGAGAAGCCAGCGAAGGGACTTCGGCAGTAGGAACTACAGAAGTCACGGAAGTGATCGACTCTGAACCCGTTTCTGAAGTTATTTGCGCGGTATGCAACTGCAGCGACAGTGTTGTGAATTGCAGTGGCAAGGAAATCCGTACCTCCTTCGAGATGACTGACTGGGAGGGTCTGCAAGAATTCAAACCAGTTAGTGTTGACCTTAGCAATAATCACATAGGTACGTTAACGAGAATCTCAAAATTGGATACCTTGAAATACTTGAATGTGTCAAACTGCGAACTTCTCGGCATCGAACATGCGACCTTCAGTTATCTACAGGAATTAGTTAGTTTGGATTTAAGCAGGAATCAATTGTTAAGTTCCTCAATCAACAAAAGGGTGTTTGTGGGTATTATAGATCCGATCCTAGGTCTTCTACCTTTCTCGAAACTCCGGTATTTAAGTTTGGCCAACAACGAAATCCATTCGCTACCACAAGACGTGTTCCTGTTTATGAATGAATTGACAACCCTTGATTTAAGTGACAATCCTATGGCGTATATCGATCAAGTAACAATGGGAGCTATCACCGATTTGGACGACCTGCAGGAACTCTACCTTCGGGGCTGTGAATTAGAAAGTCTTCCGGAAGGTTTCTTTAGAAGGCAGCGTCGTCTCGTGAAGCTGGATCTGAGCAATAACAGATTCACCACCGTTCCTGCCGTACTGGCAGAAGCAGTCACTCTGAAGTATCTGAACTTTGGCAGGAATCCTGTTGGAGCGCTGAACGATACCTCGGTATTTCCAAATCTGATTAGGCTGCGAGAGCTCCGCCTATGTCGCATGAGTAAGCTGCGCTCAGTGTCTGCCCGCGCTCTCGGAGGTCTCAAGAGTCTGGAAACATTACATTTGTGCTCGAATCCACGTCTCACCGATTTGGATCCTGAATTCTTGACGTGGACGGAGGATGAAGTAGAAATGTGGCCGACTTTGAGAGAGCTGCGTCTCAACAACAATAATCTATCGACAATCCACTCGAATTACCTAGCCCGCTGGGACCGGCTGAACGCCTTAGACTTCGGCAACAACCCCTACATCTGTGACTGCAGGAGTCAATGGACTATTGACGTGCTCATACCAATCATTCTCAAAAGTTCCCATAACGAAACTGCCAGTCATATGATCTGTAAAATGCCTCACGAAACCAGAGGTCTAACCCTGGTCCAGTTGTTTGAGACATCGAAGAAGCTCTCTTGCCTTGAAAACGAGAGCCTCAAGGCATCCCCTGGGCCGGACATGGCTATTCTACTTGGAATCATGATTGGTATATTCGTAACATTCCCTATAGTTCTGATAGTAATCTTGCTGTGGAGGAGAGGGTTCTTTGCGCGTTGCCGTTCGAAAACCATTGAGAAGGATGAAGATGAGGAATCGTATCCGCTCTGA
Protein
MFYCKYMFIILFLLTRTQKQISQAAELQAIEQTGTVTEIIPSVTVSALADFESNGETTTDVAVLKQGEASEGTSAVGTTEVTEVIDSEPVSEVICAVCNCSDSVVNCSGKEIRTSFEMTDWEGLQEFKPVSVDLSNNHIGTLTRISKLDTLKYLNVSNCELLGIEHATFSYLQELVSLDLSRNQLLSSSINKRVFVGIIDPILGLLPFSKLRYLSLANNEIHSLPQDVFLFMNELTTLDLSDNPMAYIDQVTMGAITDLDDLQELYLRGCELESLPEGFFRRQRRLVKLDLSNNRFTTVPAVLAEAVTLKYLNFGRNPVGALNDTSVFPNLIRLRELRLCRMSKLRSVSARALGGLKSLETLHLCSNPRLTDLDPEFLTWTEDEVEMWPTLRELRLNNNNLSTIHSNYLARWDRLNALDFGNNPYICDCRSQWTIDVLIPIILKSSHNETASHMICKMPHETRGLTLVQLFETSKKLSCLENESLKASPGPDMAILLGIMIGIFVTFPIVLIVILLWRRGFFARCRSKTIEKDEDEESYPL
Summary
Uniprot
H9JNJ6
A0A2A4JLS1
A0A2W1BMS2
A0A2A4JL00
A0A2W1BKI3
A0A2H1WHS9
+ More
A0A194PTP7 H9JNJ5 A0A194RN09 A0A3S2NVG7 E2BHI9 A0A0C9QDK7 A0A1W4WJZ5 A0A088AD10 A0A158NHH8 A0A2A3EK65 A0A154PES5 A0A195AVC3 F4WVP7 A0A0M8ZUW4 A0A067R5N2 A0A2P8XGE3 A0A195D4C5 A0A026WSV6 K7ISR8 E2AJI2 A0A2J7Q0W9 A0A151XFB0 A0A182H3K1 A0A023EUJ3 A0A232F0N9 A0A182JY92 A0A182SUI7 U5EN60 A0A182P5U0 Q17IL4 A0A195E2E8 A0A182QLN0 Q7Q0F7 A0A182RD26 A0A182PL02 A0A182Y1F1 D2A4M8 A0A195FYI4 A0A182MAA0 A0A0L7KT86 A0A0L7QWZ7 A0A1B0GJL0 A0A1B6JDB4 A0A0J7NFK4 A0A1Y1LJ54 A0A1L8D9V6 A0A1L8DA95 A0A182U3V5 E0W140 A0A1Q3FM23 A0A1L8DAA7 A0A1L8D9X1 A0A1Q3FMW8 A0A1Q3FN08 T1IG52 A0A0P4VPE2 A0A182KS95 R4G3L2 A0A023EQP1 V5G361 Q17IL3 A0A2S2QBU6 A0A224XBY4 R4WI11 E9I858 A0A2M4BK90 A0A2M4BJW6 T1EA71 A0A182FBB6 A0A0L0BY65 A0A2M4AQC4 A0A0K8UP11 A0A182XF02 A0A182WEW0 A0A034W330 A0A034W5S6 W5JFS7 W8B4R2 A0A084WIB3 W8BS49 A0A2H8TM79 J3JT89 N6U3U4 U4ULG7 A0A2M4BMP6 A0A1J1IH94 A0A182JB39 A0A0C9RCV0
A0A194PTP7 H9JNJ5 A0A194RN09 A0A3S2NVG7 E2BHI9 A0A0C9QDK7 A0A1W4WJZ5 A0A088AD10 A0A158NHH8 A0A2A3EK65 A0A154PES5 A0A195AVC3 F4WVP7 A0A0M8ZUW4 A0A067R5N2 A0A2P8XGE3 A0A195D4C5 A0A026WSV6 K7ISR8 E2AJI2 A0A2J7Q0W9 A0A151XFB0 A0A182H3K1 A0A023EUJ3 A0A232F0N9 A0A182JY92 A0A182SUI7 U5EN60 A0A182P5U0 Q17IL4 A0A195E2E8 A0A182QLN0 Q7Q0F7 A0A182RD26 A0A182PL02 A0A182Y1F1 D2A4M8 A0A195FYI4 A0A182MAA0 A0A0L7KT86 A0A0L7QWZ7 A0A1B0GJL0 A0A1B6JDB4 A0A0J7NFK4 A0A1Y1LJ54 A0A1L8D9V6 A0A1L8DA95 A0A182U3V5 E0W140 A0A1Q3FM23 A0A1L8DAA7 A0A1L8D9X1 A0A1Q3FMW8 A0A1Q3FN08 T1IG52 A0A0P4VPE2 A0A182KS95 R4G3L2 A0A023EQP1 V5G361 Q17IL3 A0A2S2QBU6 A0A224XBY4 R4WI11 E9I858 A0A2M4BK90 A0A2M4BJW6 T1EA71 A0A182FBB6 A0A0L0BY65 A0A2M4AQC4 A0A0K8UP11 A0A182XF02 A0A182WEW0 A0A034W330 A0A034W5S6 W5JFS7 W8B4R2 A0A084WIB3 W8BS49 A0A2H8TM79 J3JT89 N6U3U4 U4ULG7 A0A2M4BMP6 A0A1J1IH94 A0A182JB39 A0A0C9RCV0
Pubmed
19121390
28756777
26354079
20798317
21347285
21719571
+ More
24845553 29403074 24508170 30249741 20075255 26483478 24945155 28648823 17510324 12364791 25244985 18362917 19820115 26227816 28004739 20566863 27129103 20966253 23691247 21282665 26108605 25348373 20920257 23761445 24495485 24438588 22516182 23537049
24845553 29403074 24508170 30249741 20075255 26483478 24945155 28648823 17510324 12364791 25244985 18362917 19820115 26227816 28004739 20566863 27129103 20966253 23691247 21282665 26108605 25348373 20920257 23761445 24495485 24438588 22516182 23537049
EMBL
BABH01041258
NWSH01001078
PCG72726.1
KZ149991
PZC75591.1
PCG72727.1
+ More
PZC75592.1 ODYU01008753 SOQ52625.1 KQ459593 KPI96348.1 KQ460152 KPJ17386.1 RSAL01000156 RVE45598.1 GL448301 EFN84842.1 GBYB01001389 JAG71156.1 ADTU01015829 ADTU01015830 KZ288220 PBC32165.1 KQ434889 KZC10349.1 KQ976737 KYM75924.1 GL888393 EGI61717.1 KQ435878 KOX70019.1 KK852945 KDR13449.1 PYGN01002234 PSN31055.1 KQ976870 KYN07732.1 KK107131 QOIP01000012 EZA58169.1 RLU15948.1 AAZX01002901 GL440027 EFN66377.1 NEVH01019960 PNF22240.1 KQ982194 KYQ59047.1 JXUM01025059 JXUM01025060 KQ560709 KXJ81202.1 GAPW01000997 JAC12601.1 NNAY01001330 OXU24336.1 GANO01004248 JAB55623.1 CH477238 EAT46546.1 KQ979763 KYN19251.1 AXCN02001027 AAAB01008986 EAA00711.3 KQ971344 EFA05244.1 KQ981208 KYN44894.1 AXCM01014879 JTDY01005908 KOB66493.1 KQ414706 KOC63091.1 AJWK01021911 GECU01016276 GECU01010773 JAS91430.1 JAS96933.1 LBMM01005643 KMQ91340.1 GEZM01055824 JAV72921.1 GFDF01010828 JAV03256.1 GFDF01010824 JAV03260.1 DS235866 EEB19346.1 GFDL01006448 JAV28597.1 GFDF01010830 JAV03254.1 GFDF01010825 JAV03259.1 GFDL01006115 JAV28930.1 GFDL01006123 JAV28922.1 ACPB03011560 ACPB03011561 GDKW01002381 JAI54214.1 GAHY01002110 JAA75400.1 GAPW01001676 JAC11922.1 GALX01004031 JAB64435.1 EAT46547.1 GGMS01006026 MBY75229.1 GFTR01006500 JAW09926.1 AK417000 BAN20215.1 GL761471 EFZ23241.1 GGFJ01004230 MBW53371.1 GGFJ01004228 MBW53369.1 GAMD01000692 JAB00899.1 JRES01001167 KNC24911.1 GGFK01009487 MBW42808.1 GDHF01023892 JAI28422.1 GAKP01009833 JAC49119.1 GAKP01009834 JAC49118.1 ADMH02001279 ETN63232.1 GAMC01014502 JAB92053.1 ATLV01023925 KE525347 KFB49957.1 GAMC01014501 GAMC01014499 JAB92054.1 GFXV01003458 MBW15263.1 BT126445 AEE61409.1 APGK01050579 APGK01050580 APGK01050581 KB741174 ENN73247.1 KB632260 ERL90835.1 GGFJ01005184 MBW54325.1 CVRI01000051 CRK99621.1 GBYB01010962 JAG80729.1
PZC75592.1 ODYU01008753 SOQ52625.1 KQ459593 KPI96348.1 KQ460152 KPJ17386.1 RSAL01000156 RVE45598.1 GL448301 EFN84842.1 GBYB01001389 JAG71156.1 ADTU01015829 ADTU01015830 KZ288220 PBC32165.1 KQ434889 KZC10349.1 KQ976737 KYM75924.1 GL888393 EGI61717.1 KQ435878 KOX70019.1 KK852945 KDR13449.1 PYGN01002234 PSN31055.1 KQ976870 KYN07732.1 KK107131 QOIP01000012 EZA58169.1 RLU15948.1 AAZX01002901 GL440027 EFN66377.1 NEVH01019960 PNF22240.1 KQ982194 KYQ59047.1 JXUM01025059 JXUM01025060 KQ560709 KXJ81202.1 GAPW01000997 JAC12601.1 NNAY01001330 OXU24336.1 GANO01004248 JAB55623.1 CH477238 EAT46546.1 KQ979763 KYN19251.1 AXCN02001027 AAAB01008986 EAA00711.3 KQ971344 EFA05244.1 KQ981208 KYN44894.1 AXCM01014879 JTDY01005908 KOB66493.1 KQ414706 KOC63091.1 AJWK01021911 GECU01016276 GECU01010773 JAS91430.1 JAS96933.1 LBMM01005643 KMQ91340.1 GEZM01055824 JAV72921.1 GFDF01010828 JAV03256.1 GFDF01010824 JAV03260.1 DS235866 EEB19346.1 GFDL01006448 JAV28597.1 GFDF01010830 JAV03254.1 GFDF01010825 JAV03259.1 GFDL01006115 JAV28930.1 GFDL01006123 JAV28922.1 ACPB03011560 ACPB03011561 GDKW01002381 JAI54214.1 GAHY01002110 JAA75400.1 GAPW01001676 JAC11922.1 GALX01004031 JAB64435.1 EAT46547.1 GGMS01006026 MBY75229.1 GFTR01006500 JAW09926.1 AK417000 BAN20215.1 GL761471 EFZ23241.1 GGFJ01004230 MBW53371.1 GGFJ01004228 MBW53369.1 GAMD01000692 JAB00899.1 JRES01001167 KNC24911.1 GGFK01009487 MBW42808.1 GDHF01023892 JAI28422.1 GAKP01009833 JAC49119.1 GAKP01009834 JAC49118.1 ADMH02001279 ETN63232.1 GAMC01014502 JAB92053.1 ATLV01023925 KE525347 KFB49957.1 GAMC01014501 GAMC01014499 JAB92054.1 GFXV01003458 MBW15263.1 BT126445 AEE61409.1 APGK01050579 APGK01050580 APGK01050581 KB741174 ENN73247.1 KB632260 ERL90835.1 GGFJ01005184 MBW54325.1 CVRI01000051 CRK99621.1 GBYB01010962 JAG80729.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000008237
+ More
UP000192223 UP000005203 UP000005205 UP000242457 UP000076502 UP000078540 UP000007755 UP000053105 UP000027135 UP000245037 UP000078542 UP000053097 UP000279307 UP000002358 UP000000311 UP000235965 UP000075809 UP000069940 UP000249989 UP000215335 UP000075881 UP000075901 UP000075885 UP000008820 UP000078492 UP000075886 UP000007062 UP000075900 UP000076408 UP000007266 UP000078541 UP000075883 UP000037510 UP000053825 UP000092461 UP000036403 UP000075902 UP000009046 UP000015103 UP000075882 UP000069272 UP000037069 UP000076407 UP000075920 UP000000673 UP000030765 UP000019118 UP000030742 UP000183832 UP000075880
UP000192223 UP000005203 UP000005205 UP000242457 UP000076502 UP000078540 UP000007755 UP000053105 UP000027135 UP000245037 UP000078542 UP000053097 UP000279307 UP000002358 UP000000311 UP000235965 UP000075809 UP000069940 UP000249989 UP000215335 UP000075881 UP000075901 UP000075885 UP000008820 UP000078492 UP000075886 UP000007062 UP000075900 UP000076408 UP000007266 UP000078541 UP000075883 UP000037510 UP000053825 UP000092461 UP000036403 UP000075902 UP000009046 UP000015103 UP000075882 UP000069272 UP000037069 UP000076407 UP000075920 UP000000673 UP000030765 UP000019118 UP000030742 UP000183832 UP000075880
Interpro
SUPFAM
SSF46689
SSF46689
Gene 3D
ProteinModelPortal
H9JNJ6
A0A2A4JLS1
A0A2W1BMS2
A0A2A4JL00
A0A2W1BKI3
A0A2H1WHS9
+ More
A0A194PTP7 H9JNJ5 A0A194RN09 A0A3S2NVG7 E2BHI9 A0A0C9QDK7 A0A1W4WJZ5 A0A088AD10 A0A158NHH8 A0A2A3EK65 A0A154PES5 A0A195AVC3 F4WVP7 A0A0M8ZUW4 A0A067R5N2 A0A2P8XGE3 A0A195D4C5 A0A026WSV6 K7ISR8 E2AJI2 A0A2J7Q0W9 A0A151XFB0 A0A182H3K1 A0A023EUJ3 A0A232F0N9 A0A182JY92 A0A182SUI7 U5EN60 A0A182P5U0 Q17IL4 A0A195E2E8 A0A182QLN0 Q7Q0F7 A0A182RD26 A0A182PL02 A0A182Y1F1 D2A4M8 A0A195FYI4 A0A182MAA0 A0A0L7KT86 A0A0L7QWZ7 A0A1B0GJL0 A0A1B6JDB4 A0A0J7NFK4 A0A1Y1LJ54 A0A1L8D9V6 A0A1L8DA95 A0A182U3V5 E0W140 A0A1Q3FM23 A0A1L8DAA7 A0A1L8D9X1 A0A1Q3FMW8 A0A1Q3FN08 T1IG52 A0A0P4VPE2 A0A182KS95 R4G3L2 A0A023EQP1 V5G361 Q17IL3 A0A2S2QBU6 A0A224XBY4 R4WI11 E9I858 A0A2M4BK90 A0A2M4BJW6 T1EA71 A0A182FBB6 A0A0L0BY65 A0A2M4AQC4 A0A0K8UP11 A0A182XF02 A0A182WEW0 A0A034W330 A0A034W5S6 W5JFS7 W8B4R2 A0A084WIB3 W8BS49 A0A2H8TM79 J3JT89 N6U3U4 U4ULG7 A0A2M4BMP6 A0A1J1IH94 A0A182JB39 A0A0C9RCV0
A0A194PTP7 H9JNJ5 A0A194RN09 A0A3S2NVG7 E2BHI9 A0A0C9QDK7 A0A1W4WJZ5 A0A088AD10 A0A158NHH8 A0A2A3EK65 A0A154PES5 A0A195AVC3 F4WVP7 A0A0M8ZUW4 A0A067R5N2 A0A2P8XGE3 A0A195D4C5 A0A026WSV6 K7ISR8 E2AJI2 A0A2J7Q0W9 A0A151XFB0 A0A182H3K1 A0A023EUJ3 A0A232F0N9 A0A182JY92 A0A182SUI7 U5EN60 A0A182P5U0 Q17IL4 A0A195E2E8 A0A182QLN0 Q7Q0F7 A0A182RD26 A0A182PL02 A0A182Y1F1 D2A4M8 A0A195FYI4 A0A182MAA0 A0A0L7KT86 A0A0L7QWZ7 A0A1B0GJL0 A0A1B6JDB4 A0A0J7NFK4 A0A1Y1LJ54 A0A1L8D9V6 A0A1L8DA95 A0A182U3V5 E0W140 A0A1Q3FM23 A0A1L8DAA7 A0A1L8D9X1 A0A1Q3FMW8 A0A1Q3FN08 T1IG52 A0A0P4VPE2 A0A182KS95 R4G3L2 A0A023EQP1 V5G361 Q17IL3 A0A2S2QBU6 A0A224XBY4 R4WI11 E9I858 A0A2M4BK90 A0A2M4BJW6 T1EA71 A0A182FBB6 A0A0L0BY65 A0A2M4AQC4 A0A0K8UP11 A0A182XF02 A0A182WEW0 A0A034W330 A0A034W5S6 W5JFS7 W8B4R2 A0A084WIB3 W8BS49 A0A2H8TM79 J3JT89 N6U3U4 U4ULG7 A0A2M4BMP6 A0A1J1IH94 A0A182JB39 A0A0C9RCV0
PDB
4R5D
E-value=1.13063e-19,
Score=239
Ontologies
GO
Topology
Subcellular location
Nucleus
SignalP
Position: 1 - 24,
Likelihood: 0.930888
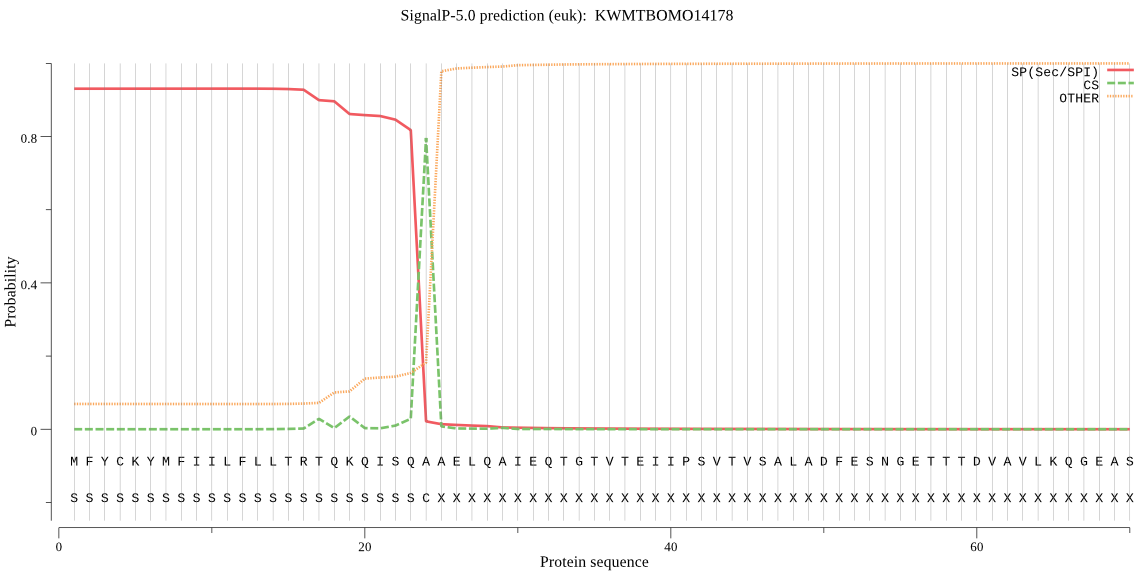
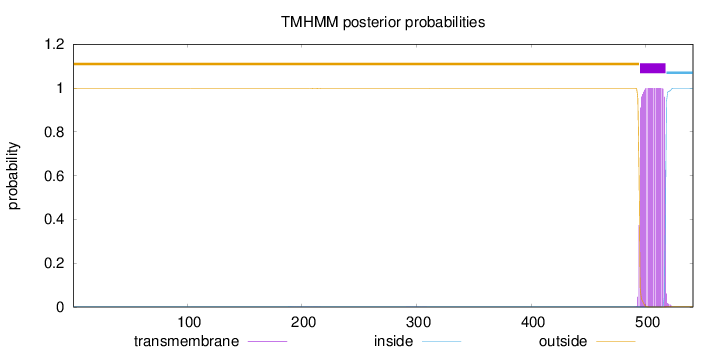
Length:
541
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.96449
Exp number, first 60 AAs:
0.00189
Total prob of N-in:
0.00213
outside
1 - 494
TMhelix
495 - 517
inside
518 - 541
Population Genetic Test Statistics
Pi
20.457234
Theta
17.459221
Tajima's D
1.132834
CLR
0.633601
CSRT
0.706314684265787
Interpretation
Uncertain
