Pre Gene Modal
BGIBMGA011170
Annotation
PREDICTED:_anoctamin-4-like_isoform_X2_[Bombyx_mori]
Full name
Anoctamin
Location in the cell
PlasmaMembrane Reliability : 4.059
Sequence
CDS
ATGTATTATTTAGCCACAACATTTCTGGAGCTCTGGAAGAGGAAGCAGTCCGTGCTGAGATGGGAGTGGGACTTGGGTGGCGTCGACTATGACGAGGACCCTCGGCCAGAGTTCGAAACCTCAGCGAAGACATTCCGCACCAACCCTGTGACCAGGGAGAAGGAGCCTTATCTACCAGTCTGGCAGAAGACCATGAGATACATCGCGACGAGCAGCGCAGTTTTGTTCATGATCGCGATCGTGATGGGAGCGGTCCTCGGGACCATCATCTACAGGATCTCCATGGTGTCAGTCATCTACGGTGGCTCGGGGTTCTTCATACAGCGGCACGCAAAGATCTTCACCGCCATGACAGCAGCCCTGATCAATTTAATCATCATCATGATTCTGACCAGGATATACGCCAAGGTGGCGGTCTATCTGACGAACATGGAGAACCCTCGCACCCAAACGGAATACGAAGACTCGTACACCTTCAAAATATTTTTCTTTGAGTTCATGAACTTTTATTCTTCTTTGATTTACATTGCGTTCTTTAAGGGTCGTTTCTACGACTACCCCGGCGACGACCAGGCCAGGAAGTCGGAGTTCTTCAAGCTAAAAGGCGACATCTGCGATCCGGCCGGCTGTCTCTCCGAGCTGTGCATACAACTAGCCATCATTATGATTGGGAAGCAATTCTTCAACAACTTCTTGGAACTGGGATTCCCCAAGTTCCACAATTGGTGGCGTCGTCGCTCGCACCGGGCCGTGACCAGGGACCCCAGCAAGCCGCACATGGCGTGGGAACAGGACTACCACCTCCAGGACCCTGGGAGGCTGGCCTTGTTCGATGAATACCTAGAAATGATCCTCCAATACGGTTTCGTGACGTTATTTGTGGCCGCCTTCCCGCTGGCCCCACTGTTCGCATTGCTCAACAACATAGCCGAGATCAGACTGGACGCGTACAAAATGGTGACACAGGCGAGGCGACCTCTAGCGGAGAGGGTCGAGGACATCGGAGCCTGGTACGGGATATTGCGTGGGATAACGTATGCGGCGGTCGTCTCAAATGCGTTCGTGATAGCGTACACAAGTGATTTCATTCCGCGTATGGTCTACAAATACGTCTATTCCCCGGACAACACCCTAGCAGGATACATAGACCATTCGTTGTCTCTGTTCAACACATCAGATTACCGGGAAGACTGGGGGGCTGACGAGAACGAAGTTGACCCCCCGGTGTGTGCGTACCGCGGGTACCGGAATCCGCCAGACCACGCAGACCCCTACGGTCTGTCTCCGCACTACTGGCACGTGTTCGCGGCAAGACTGGCCTTCGTCGTCGTATTCGAGCATGTTGTATTCGGTCTAACGGGCCTAATGCAGCTGTTTATACCCGATTTGCCGAGCGAACTGCGGATACAGATGCAAAGGGAAGCGCTGCTAGCTAAGGAAGCGAAATATGAGCACGGCAGAAGAAGATTGTCTACCGAATACAGACAGCTGATCACGCAGAAAAGCGAGAGCAGCGGAGAGCGCCCCGCTGGTCCGTGGCTGCGTCGGACCTCCCGGGCCTCGGACGGGCTGGACGCGCACGTTGCCGTCGCCCAGAAACCTTCTTCTCCACAAATAACGCACTGA
Protein
MYYLATTFLELWKRKQSVLRWEWDLGGVDYDEDPRPEFETSAKTFRTNPVTREKEPYLPVWQKTMRYIATSSAVLFMIAIVMGAVLGTIIYRISMVSVIYGGSGFFIQRHAKIFTAMTAALINLIIIMILTRIYAKVAVYLTNMENPRTQTEYEDSYTFKIFFFEFMNFYSSLIYIAFFKGRFYDYPGDDQARKSEFFKLKGDICDPAGCLSELCIQLAIIMIGKQFFNNFLELGFPKFHNWWRRRSHRAVTRDPSKPHMAWEQDYHLQDPGRLALFDEYLEMILQYGFVTLFVAAFPLAPLFALLNNIAEIRLDAYKMVTQARRPLAERVEDIGAWYGILRGITYAAVVSNAFVIAYTSDFIPRMVYKYVYSPDNTLAGYIDHSLSLFNTSDYREDWGADENEVDPPVCAYRGYRNPPDHADPYGLSPHYWHVFAARLAFVVVFEHVVFGLTGLMQLFIPDLPSELRIQMQREALLAKEAKYEHGRRRLSTEYRQLITQKSESSGERPAGPWLRRTSRASDGLDAHVAVAQKPSSPQITH
Summary
Similarity
Belongs to the anoctamin family.
Feature
chain Anoctamin
Uniprot
H9JNR6
A0A2A4K343
A0A3S2LAU8
A0A2W1BSG4
A0A194PZ22
A0A212FPR5
+ More
A0A0N0PDV9 A0A182QPT7 A0A084VUQ2 Q0IEX5 B4J0P8 A0A1S4FHG8 B4LHU1 B4L017 A0A1S4H4Y1 Q7QDY0 A0A182YIT8 W5JG03 W8CA60 A0A1W4W677 Q9VTS0 M9PI24 B3NH46 B4PG75 B4MKX3 A0A0J9RTR1 B4QQR6 B3M4N6 A0A3B0KHV6 A0A182U8K8 Q2M0Y5 A0A182IPR9 A0A1I8MV46 A0A2H1WLT2 A0A182X1N3 B4GQY0 A0A1I8PA10 A0A1B0B1R2 A0A1A9WRI9 A0A1A9XYW5 A0A1A9UQQ3 A0A336M0R5 A0A1B0G917 D2A4M0 N6U0P0 A0A1J1IWQ2 A0A1A9Z2X2 A0A1B0CV41 A0A0L7R9Y8 A0A182FBV7 A0A310SNK3 A0A232F783 A0A182W3X2 A0A182RJT0 A0A182HXE0 K7J4Y8 A0A182PLU2 A0A2A3EMB4 E0VCR5 A0A154PEI2 A0A0C9REX8 A0A026W6U2 E2BCH4 A0A088AD01 A0A0K8SA94 A0A0K8SBQ9 A0A0K8SA97 A0A0K8SKC7 E9IY00 A0A182JV28 E2AZ67 A0A182NDF6 A0A195DCM7 A0A151XA31 A0A195FW00 A0A158P3V6 A0A0L0BVU8 A0A151IGH3 A0A3L8DY79 F4WM46 T1HSW8 A0A067RA79 A0A3Q0IJZ5 A0A2H1V5K3 A0A226E438 R7UV80 T1J757 A0A091FNW0 F1M4U7 A0A1A8LC53 A0A093RYU4 A0A091T171 A0A093IME6 A0A093BMI9 A0A1A8RH66 A0A1A8BCZ1 U3K996 A0A091QWS4 A0A1A8I3I1
A0A0N0PDV9 A0A182QPT7 A0A084VUQ2 Q0IEX5 B4J0P8 A0A1S4FHG8 B4LHU1 B4L017 A0A1S4H4Y1 Q7QDY0 A0A182YIT8 W5JG03 W8CA60 A0A1W4W677 Q9VTS0 M9PI24 B3NH46 B4PG75 B4MKX3 A0A0J9RTR1 B4QQR6 B3M4N6 A0A3B0KHV6 A0A182U8K8 Q2M0Y5 A0A182IPR9 A0A1I8MV46 A0A2H1WLT2 A0A182X1N3 B4GQY0 A0A1I8PA10 A0A1B0B1R2 A0A1A9WRI9 A0A1A9XYW5 A0A1A9UQQ3 A0A336M0R5 A0A1B0G917 D2A4M0 N6U0P0 A0A1J1IWQ2 A0A1A9Z2X2 A0A1B0CV41 A0A0L7R9Y8 A0A182FBV7 A0A310SNK3 A0A232F783 A0A182W3X2 A0A182RJT0 A0A182HXE0 K7J4Y8 A0A182PLU2 A0A2A3EMB4 E0VCR5 A0A154PEI2 A0A0C9REX8 A0A026W6U2 E2BCH4 A0A088AD01 A0A0K8SA94 A0A0K8SBQ9 A0A0K8SA97 A0A0K8SKC7 E9IY00 A0A182JV28 E2AZ67 A0A182NDF6 A0A195DCM7 A0A151XA31 A0A195FW00 A0A158P3V6 A0A0L0BVU8 A0A151IGH3 A0A3L8DY79 F4WM46 T1HSW8 A0A067RA79 A0A3Q0IJZ5 A0A2H1V5K3 A0A226E438 R7UV80 T1J757 A0A091FNW0 F1M4U7 A0A1A8LC53 A0A093RYU4 A0A091T171 A0A093IME6 A0A093BMI9 A0A1A8RH66 A0A1A8BCZ1 U3K996 A0A091QWS4 A0A1A8I3I1
Pubmed
19121390
28756777
26354079
22118469
24438588
17510324
+ More
17994087 12364791 25244985 20920257 23761445 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 15632085 25315136 18362917 19820115 23537049 28648823 20075255 20566863 24508170 20798317 21282665 21347285 26108605 30249741 21719571 24845553 23254933 15057822 15632090
17994087 12364791 25244985 20920257 23761445 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 15632085 25315136 18362917 19820115 23537049 28648823 20075255 20566863 24508170 20798317 21282665 21347285 26108605 30249741 21719571 24845553 23254933 15057822 15632090
EMBL
BABH01010041
BABH01010042
NWSH01000181
PCG78681.1
RSAL01000062
RVE49577.1
+ More
KZ149991 PZC75586.1 KQ459593 KPI96405.1 AGBW02001601 OWR55699.1 KQ460036 KPJ18428.1 AXCN02002125 ATLV01016983 ATLV01016984 ATLV01016985 ATLV01016986 KE525139 KFB41696.1 CH477461 EAT40566.1 CH916366 EDV97903.1 CH940647 EDW70666.2 CH933809 EDW19052.2 AAAB01008849 EAA07133.4 ADMH02001609 ETN61810.1 GAMC01001559 JAC04997.1 AE014296 AAF49976.2 AGB94426.1 CH954178 EDV51503.2 CM000159 EDW94239.2 CH963847 EDW72898.2 CM002912 KMY99128.1 CM000363 EDX10168.1 CH902618 EDV39435.2 OUUW01000009 SPP84691.1 CH379069 EAL30791.3 ODYU01009471 SOQ53936.1 CH479188 EDW40165.1 JXJN01007264 UFQT01000292 SSX22911.1 CCAG010002961 KQ971344 EFA05242.1 APGK01044219 KB741021 ENN75070.1 CVRI01000059 CRL03590.1 AJWK01030095 AJWK01030096 KQ414620 KOC67687.1 KQ759773 OAD62918.1 NNAY01000744 OXU26716.1 APCN01005189 KZ288220 PBC32181.1 DS235063 EEB11171.1 KQ434889 KZC10272.1 GBYB01005541 JAG75308.1 KK107399 EZA51346.1 GL447283 EFN86600.1 GBRD01015619 JAG50207.1 GBRD01015621 GBRD01012041 JAG50205.1 GBRD01015620 GBRD01012043 JAG50206.1 GBRD01012042 JAG53782.1 GL766836 EFZ14563.1 GL444096 EFN61274.1 KQ980989 KYN10592.1 KQ982353 KYQ57213.1 KQ981264 KYN44019.1 ADTU01001341 JRES01001255 KNC24162.1 KQ977726 KYN00321.1 QOIP01000002 RLU25404.1 GL888217 EGI64690.1 ACPB03016662 KK852602 KDR20488.1 ODYU01000790 SOQ36107.1 LNIX01000006 OXA52495.1 AMQN01007128 KB300038 ELU07311.1 JH431912 KL447189 KFO70676.1 AABR07056545 AABR07056546 AABR07056547 AABR07056548 AABR07056549 AABR07056550 AABR07056551 CH473960 EDM16991.1 HAEF01004701 SBR42083.1 KL413618 KFW75914.1 KK495622 KFQ65404.1 KK590763 KFV99928.1 KN126564 KFU90665.1 HAEH01016998 SBS05430.1 HADZ01001481 SBP65422.1 AGTO01000061 AGTO01020300 KK708706 KFQ32025.1 HAED01005440 SBQ91470.1
KZ149991 PZC75586.1 KQ459593 KPI96405.1 AGBW02001601 OWR55699.1 KQ460036 KPJ18428.1 AXCN02002125 ATLV01016983 ATLV01016984 ATLV01016985 ATLV01016986 KE525139 KFB41696.1 CH477461 EAT40566.1 CH916366 EDV97903.1 CH940647 EDW70666.2 CH933809 EDW19052.2 AAAB01008849 EAA07133.4 ADMH02001609 ETN61810.1 GAMC01001559 JAC04997.1 AE014296 AAF49976.2 AGB94426.1 CH954178 EDV51503.2 CM000159 EDW94239.2 CH963847 EDW72898.2 CM002912 KMY99128.1 CM000363 EDX10168.1 CH902618 EDV39435.2 OUUW01000009 SPP84691.1 CH379069 EAL30791.3 ODYU01009471 SOQ53936.1 CH479188 EDW40165.1 JXJN01007264 UFQT01000292 SSX22911.1 CCAG010002961 KQ971344 EFA05242.1 APGK01044219 KB741021 ENN75070.1 CVRI01000059 CRL03590.1 AJWK01030095 AJWK01030096 KQ414620 KOC67687.1 KQ759773 OAD62918.1 NNAY01000744 OXU26716.1 APCN01005189 KZ288220 PBC32181.1 DS235063 EEB11171.1 KQ434889 KZC10272.1 GBYB01005541 JAG75308.1 KK107399 EZA51346.1 GL447283 EFN86600.1 GBRD01015619 JAG50207.1 GBRD01015621 GBRD01012041 JAG50205.1 GBRD01015620 GBRD01012043 JAG50206.1 GBRD01012042 JAG53782.1 GL766836 EFZ14563.1 GL444096 EFN61274.1 KQ980989 KYN10592.1 KQ982353 KYQ57213.1 KQ981264 KYN44019.1 ADTU01001341 JRES01001255 KNC24162.1 KQ977726 KYN00321.1 QOIP01000002 RLU25404.1 GL888217 EGI64690.1 ACPB03016662 KK852602 KDR20488.1 ODYU01000790 SOQ36107.1 LNIX01000006 OXA52495.1 AMQN01007128 KB300038 ELU07311.1 JH431912 KL447189 KFO70676.1 AABR07056545 AABR07056546 AABR07056547 AABR07056548 AABR07056549 AABR07056550 AABR07056551 CH473960 EDM16991.1 HAEF01004701 SBR42083.1 KL413618 KFW75914.1 KK495622 KFQ65404.1 KK590763 KFV99928.1 KN126564 KFU90665.1 HAEH01016998 SBS05430.1 HADZ01001481 SBP65422.1 AGTO01000061 AGTO01020300 KK708706 KFQ32025.1 HAED01005440 SBQ91470.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
+ More
UP000075886 UP000030765 UP000008820 UP000001070 UP000008792 UP000009192 UP000007062 UP000076408 UP000000673 UP000192221 UP000000803 UP000008711 UP000002282 UP000007798 UP000000304 UP000007801 UP000268350 UP000075902 UP000001819 UP000075880 UP000095301 UP000076407 UP000008744 UP000095300 UP000092460 UP000091820 UP000092443 UP000078200 UP000092444 UP000007266 UP000019118 UP000183832 UP000092445 UP000092461 UP000053825 UP000069272 UP000215335 UP000075920 UP000075900 UP000075840 UP000002358 UP000075885 UP000242457 UP000009046 UP000076502 UP000053097 UP000008237 UP000005203 UP000075881 UP000000311 UP000075884 UP000078492 UP000075809 UP000078541 UP000005205 UP000037069 UP000078542 UP000279307 UP000007755 UP000015103 UP000027135 UP000079169 UP000198287 UP000014760 UP000053760 UP000002494 UP000016665
UP000075886 UP000030765 UP000008820 UP000001070 UP000008792 UP000009192 UP000007062 UP000076408 UP000000673 UP000192221 UP000000803 UP000008711 UP000002282 UP000007798 UP000000304 UP000007801 UP000268350 UP000075902 UP000001819 UP000075880 UP000095301 UP000076407 UP000008744 UP000095300 UP000092460 UP000091820 UP000092443 UP000078200 UP000092444 UP000007266 UP000019118 UP000183832 UP000092445 UP000092461 UP000053825 UP000069272 UP000215335 UP000075920 UP000075900 UP000075840 UP000002358 UP000075885 UP000242457 UP000009046 UP000076502 UP000053097 UP000008237 UP000005203 UP000075881 UP000000311 UP000075884 UP000078492 UP000075809 UP000078541 UP000005205 UP000037069 UP000078542 UP000279307 UP000007755 UP000015103 UP000027135 UP000079169 UP000198287 UP000014760 UP000053760 UP000002494 UP000016665
Interpro
Gene 3D
ProteinModelPortal
H9JNR6
A0A2A4K343
A0A3S2LAU8
A0A2W1BSG4
A0A194PZ22
A0A212FPR5
+ More
A0A0N0PDV9 A0A182QPT7 A0A084VUQ2 Q0IEX5 B4J0P8 A0A1S4FHG8 B4LHU1 B4L017 A0A1S4H4Y1 Q7QDY0 A0A182YIT8 W5JG03 W8CA60 A0A1W4W677 Q9VTS0 M9PI24 B3NH46 B4PG75 B4MKX3 A0A0J9RTR1 B4QQR6 B3M4N6 A0A3B0KHV6 A0A182U8K8 Q2M0Y5 A0A182IPR9 A0A1I8MV46 A0A2H1WLT2 A0A182X1N3 B4GQY0 A0A1I8PA10 A0A1B0B1R2 A0A1A9WRI9 A0A1A9XYW5 A0A1A9UQQ3 A0A336M0R5 A0A1B0G917 D2A4M0 N6U0P0 A0A1J1IWQ2 A0A1A9Z2X2 A0A1B0CV41 A0A0L7R9Y8 A0A182FBV7 A0A310SNK3 A0A232F783 A0A182W3X2 A0A182RJT0 A0A182HXE0 K7J4Y8 A0A182PLU2 A0A2A3EMB4 E0VCR5 A0A154PEI2 A0A0C9REX8 A0A026W6U2 E2BCH4 A0A088AD01 A0A0K8SA94 A0A0K8SBQ9 A0A0K8SA97 A0A0K8SKC7 E9IY00 A0A182JV28 E2AZ67 A0A182NDF6 A0A195DCM7 A0A151XA31 A0A195FW00 A0A158P3V6 A0A0L0BVU8 A0A151IGH3 A0A3L8DY79 F4WM46 T1HSW8 A0A067RA79 A0A3Q0IJZ5 A0A2H1V5K3 A0A226E438 R7UV80 T1J757 A0A091FNW0 F1M4U7 A0A1A8LC53 A0A093RYU4 A0A091T171 A0A093IME6 A0A093BMI9 A0A1A8RH66 A0A1A8BCZ1 U3K996 A0A091QWS4 A0A1A8I3I1
A0A0N0PDV9 A0A182QPT7 A0A084VUQ2 Q0IEX5 B4J0P8 A0A1S4FHG8 B4LHU1 B4L017 A0A1S4H4Y1 Q7QDY0 A0A182YIT8 W5JG03 W8CA60 A0A1W4W677 Q9VTS0 M9PI24 B3NH46 B4PG75 B4MKX3 A0A0J9RTR1 B4QQR6 B3M4N6 A0A3B0KHV6 A0A182U8K8 Q2M0Y5 A0A182IPR9 A0A1I8MV46 A0A2H1WLT2 A0A182X1N3 B4GQY0 A0A1I8PA10 A0A1B0B1R2 A0A1A9WRI9 A0A1A9XYW5 A0A1A9UQQ3 A0A336M0R5 A0A1B0G917 D2A4M0 N6U0P0 A0A1J1IWQ2 A0A1A9Z2X2 A0A1B0CV41 A0A0L7R9Y8 A0A182FBV7 A0A310SNK3 A0A232F783 A0A182W3X2 A0A182RJT0 A0A182HXE0 K7J4Y8 A0A182PLU2 A0A2A3EMB4 E0VCR5 A0A154PEI2 A0A0C9REX8 A0A026W6U2 E2BCH4 A0A088AD01 A0A0K8SA94 A0A0K8SBQ9 A0A0K8SA97 A0A0K8SKC7 E9IY00 A0A182JV28 E2AZ67 A0A182NDF6 A0A195DCM7 A0A151XA31 A0A195FW00 A0A158P3V6 A0A0L0BVU8 A0A151IGH3 A0A3L8DY79 F4WM46 T1HSW8 A0A067RA79 A0A3Q0IJZ5 A0A2H1V5K3 A0A226E438 R7UV80 T1J757 A0A091FNW0 F1M4U7 A0A1A8LC53 A0A093RYU4 A0A091T171 A0A093IME6 A0A093BMI9 A0A1A8RH66 A0A1A8BCZ1 U3K996 A0A091QWS4 A0A1A8I3I1
PDB
6QPI
E-value=1.09839e-90,
Score=851
Ontologies
GO
PANTHER
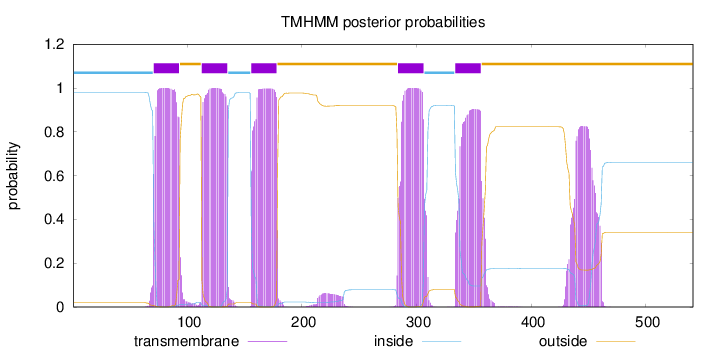
Topology
Subcellular location
Membrane
Length:
541
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
131.05479
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.97957
inside
1 - 70
TMhelix
71 - 93
outside
94 - 112
TMhelix
113 - 135
inside
136 - 155
TMhelix
156 - 178
outside
179 - 283
TMhelix
284 - 306
inside
307 - 333
TMhelix
334 - 356
outside
357 - 541
Population Genetic Test Statistics
Pi
17.770775
Theta
15.569415
Tajima's D
0.230448
CLR
0.773776
CSRT
0.43912804359782
Interpretation
Uncertain
