Gene
KWMTBOMO14167
Pre Gene Modal
BGIBMGA011415
Annotation
hypothetical_protein_KGM_02634_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 4.97
Sequence
CDS
ATGTACGGTCGAAAAACACCGACCACTTATCGTTCTACACCGTCTGTGTATTCTCATTACACAGGGAAGTCATCAACAAATCTACGCTCATCAAAGTCGGTGAGATCCCTACGAGTTCCATGGTACCAGAAGCCAATATTACATGATGCAGTCATTTTGGACCTGCAGAGAGGATCATTGCTCACTGCATTTCTTTCTTTGTTCATGTCAATTTTCACATTTGCACAAAGTGTTTTCGACTTGTACTGCCTTTACATGGCGATGCCTGGATCCCATCATACTGGTTTCTATGTTGTCAGTTATGAATTTGTCTACGTTGGAAGCCCCCCAGTACGCAATGTTTTGATAATATTTTCGCTTTTCTCATGGTTGGGTTCCATCGCCGTGTTGATAACGAGCATAATATTAATCAACGCTTTAAGGAAGGAATACGAAAAGCGAACGGTTCCCTGGCTGTACGCCTTCGGAGTTTTCATCATTTTCCGTTTCTTCGCGTTTATTTTCGCTTCGATAGTGAACGACATGATATTCGCTTACAACATCATGATGTGCTTAATATGGATAGTGTTCTGCGTGGGTGGCGTCTACGGTTGGCTGCTGGTGTACAGCCTGTATCTGGAGCTGGCTGATCTGACCAAGCTCGAAGATTTGGCGCATCTGAGGATGGGAACAATGGCGTCGTTGAATGCGTCCATAGCTGGGTCGCGACCGACGACCCCGCACAGCACGGTGTCCACGATGCCGGCTTCGCAGATATGA
Protein
MYGRKTPTTYRSTPSVYSHYTGKSSTNLRSSKSVRSLRVPWYQKPILHDAVILDLQRGSLLTAFLSLFMSIFTFAQSVFDLYCLYMAMPGSHHTGFYVVSYEFVYVGSPPVRNVLIIFSLFSWLGSIAVLITSIILINALRKEYEKRTVPWLYAFGVFIIFRFFAFIFASIVNDMIFAYNIMMCLIWIVFCVGGVYGWLLVYSLYLELADLTKLEDLAHLRMGTMASLNASIAGSRPTTPHSTVSTMPASQI
Summary
Similarity
Belongs to the MAGUK family.
Uniprot
A0A2A4JAJ9
A0A2W1BKL3
A0A2H1VY17
A0A212F049
H9JPG1
A0A194PZ27
+ More
U5EPB4 A0A067RBG4 B0WE56 A0A2P8YLX6 A0A1Q3FL93 A0A2J7RI46 A0A023ENW3 A0A182NTT8 A0A182MDE5 A0A182P8K5 A0A182Y3C9 A0A182VIB7 A0A182TII3 A0A182L0S0 A0A182XL97 Q7Q4B4 A0A182ICC7 A0A1B0D1Q8 A0A182K6F0 A0A182ISK4 A0A084VEB3 A0A1B0CTN4 A0A2M4CHD7 W5J306 A0A2M4BWN9 A0A2M4AAN6 A0A2M4AAS5 A0A2M3Z5T7 E2C5G2 A0A2M4BWG9 A0A182F1F2 E9IGP3 A0A2M4BW89 D2A3Q1 A0A154PM36 A0A0L7RBA4 A0A0C9RGT3 A0A0N1IHV2 A0A1L8DA16 A0A1W4WSR6 B3P1R3 A0A026WQX1 B4PUM0 E2AXL6 A0A1W4W1S9 Q16UD3 Q9VHF5 B4HKF4 B4QX74 A0A2A3E1A7 A0A310SCY0 Q29BL1 A0A3B0JK44 B3M072 B4NAJ3 B4K4Y4 A0A195D7U5 B4JYT1 A0A182QZK6 A0A1B6MGJ6 A0A195C523 A0A182RMB3 A0A0M5J7B9 A0A195ER62 B4LW07 A0A232ERV8 A0A1B6CNG7 A0A151X476 A0A195BR62 A0A158NDU4 A0A0A1XMI5 A0A0K8U7J1 A0A1A9Y2K8 A0A1B0B8F0 A0A034W4M5 A0A1I8NR35 A0A1I8MNF0 A0A0L0CE03 A0A1B0FAI5 A0A1A9ZUP4 A0A1A9VGF8 A0A0T6B0Z8 A0A069DQQ4 J3JT76 A0A224XW29 A0A0K8TPZ5 A0A023F567 A0A2S2R6P6 A0A0V0GA21 A0A2S2PTX1 A0A1J1I6N0 J9JKI0 A0A146L4T8 A0A1Y1NGX3
U5EPB4 A0A067RBG4 B0WE56 A0A2P8YLX6 A0A1Q3FL93 A0A2J7RI46 A0A023ENW3 A0A182NTT8 A0A182MDE5 A0A182P8K5 A0A182Y3C9 A0A182VIB7 A0A182TII3 A0A182L0S0 A0A182XL97 Q7Q4B4 A0A182ICC7 A0A1B0D1Q8 A0A182K6F0 A0A182ISK4 A0A084VEB3 A0A1B0CTN4 A0A2M4CHD7 W5J306 A0A2M4BWN9 A0A2M4AAN6 A0A2M4AAS5 A0A2M3Z5T7 E2C5G2 A0A2M4BWG9 A0A182F1F2 E9IGP3 A0A2M4BW89 D2A3Q1 A0A154PM36 A0A0L7RBA4 A0A0C9RGT3 A0A0N1IHV2 A0A1L8DA16 A0A1W4WSR6 B3P1R3 A0A026WQX1 B4PUM0 E2AXL6 A0A1W4W1S9 Q16UD3 Q9VHF5 B4HKF4 B4QX74 A0A2A3E1A7 A0A310SCY0 Q29BL1 A0A3B0JK44 B3M072 B4NAJ3 B4K4Y4 A0A195D7U5 B4JYT1 A0A182QZK6 A0A1B6MGJ6 A0A195C523 A0A182RMB3 A0A0M5J7B9 A0A195ER62 B4LW07 A0A232ERV8 A0A1B6CNG7 A0A151X476 A0A195BR62 A0A158NDU4 A0A0A1XMI5 A0A0K8U7J1 A0A1A9Y2K8 A0A1B0B8F0 A0A034W4M5 A0A1I8NR35 A0A1I8MNF0 A0A0L0CE03 A0A1B0FAI5 A0A1A9ZUP4 A0A1A9VGF8 A0A0T6B0Z8 A0A069DQQ4 J3JT76 A0A224XW29 A0A0K8TPZ5 A0A023F567 A0A2S2R6P6 A0A0V0GA21 A0A2S2PTX1 A0A1J1I6N0 J9JKI0 A0A146L4T8 A0A1Y1NGX3
Pubmed
28756777
22118469
19121390
26354079
24845553
29403074
+ More
24945155 26483478 25244985 20966253 12364791 14747013 17210077 24438588 20920257 23761445 20798317 21282665 18362917 19820115 17994087 24508170 30249741 17550304 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 28648823 21347285 25830018 25348373 25315136 26108605 26334808 22516182 23537049 26369729 25474469 26823975 28004739
24945155 26483478 25244985 20966253 12364791 14747013 17210077 24438588 20920257 23761445 20798317 21282665 18362917 19820115 17994087 24508170 30249741 17550304 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 28648823 21347285 25830018 25348373 25315136 26108605 26334808 22516182 23537049 26369729 25474469 26823975 28004739
EMBL
NWSH01002113
PCG69137.1
KZ149991
PZC75579.1
ODYU01005153
SOQ45731.1
+ More
AGBW02011174 OWR47102.1 BABH01010029 KQ459593 KPI96410.1 GANO01004772 JAB55099.1 KK852809 KDR16002.1 DS231904 EDS45232.1 PYGN01000502 PSN45250.1 GFDL01006708 JAV28337.1 NEVH01003505 PNF40508.1 JXUM01083118 GAPW01003459 KQ563357 JAC10139.1 KXJ73988.1 AXCM01002898 AAAB01008964 EAA12430.3 APCN01000794 AJVK01022250 AJVK01022251 ATLV01012266 KE524777 KFB36307.1 AJWK01027789 GGFL01000579 MBW64757.1 ADMH02002157 ETN58206.1 GGFJ01008346 MBW57487.1 GGFK01004530 MBW37851.1 GGFK01004554 MBW37875.1 GGFM01003131 MBW23882.1 GL452770 EFN76775.1 GGFJ01008213 MBW57354.1 GL763054 EFZ20220.1 GGFJ01008121 MBW57262.1 KQ971348 EFA05545.1 KQ434977 KZC12941.1 KQ414617 KOC68026.1 GBYB01007560 JAG77327.1 KQ460036 KPJ18422.1 GFDF01010879 JAV03205.1 CH954181 EDV49801.1 KK107126 QOIP01000008 EZA58442.1 RLU19582.1 CM000160 EDW96637.1 GL443590 EFN61854.1 CH477624 EAT38135.1 AE014297 AY051956 AAF54359.1 AAK93380.1 AAN13415.1 AAN13416.1 CH480815 EDW42904.1 CM000364 EDX13645.1 KZ288460 PBC25480.1 KQ776822 OAD52167.1 CM000070 EAL26986.2 OUUW01000005 SPP81163.1 CH902617 EDV42029.1 CH964232 EDW80807.1 CH933806 EDW13955.1 KQ981153 KYN08938.1 CH916378 EDV98546.1 AXCN02000805 GEBQ01012409 GEBQ01004952 JAT27568.1 JAT35025.1 KQ978251 KYM95967.1 CP012526 ALC47131.1 KQ981993 KYN30755.1 CH940650 EDW66512.1 NNAY01002542 OXU21098.1 GEDC01022271 JAS15027.1 KQ982548 KYQ55245.1 KQ976424 KYM88449.1 ADTU01013021 GBXI01002081 GBXI01001770 JAD12211.1 JAD12522.1 GDHF01030024 GDHF01011138 JAI22290.1 JAI41176.1 JXJN01009919 GAKP01009286 GAKP01009285 GAKP01009284 GAKP01009283 GAKP01009282 GAKP01009281 JAC49669.1 JRES01000501 KNC30643.1 CCAG010015282 LJIG01016302 KRT81010.1 GBGD01002659 JAC86230.1 APGK01054668 BT126432 KB741251 KB632404 AEE61396.1 ENN71860.1 ERL95014.1 GFTR01004103 JAW12323.1 GDAI01001136 JAI16467.1 GBBI01002498 JAC16214.1 GGMS01016167 MBY85370.1 GECL01001262 JAP04862.1 GGMR01020312 MBY32931.1 CVRI01000043 CRK95967.1 ABLF02017312 GDHC01015974 JAQ02655.1 GEZM01007120 JAV95366.1
AGBW02011174 OWR47102.1 BABH01010029 KQ459593 KPI96410.1 GANO01004772 JAB55099.1 KK852809 KDR16002.1 DS231904 EDS45232.1 PYGN01000502 PSN45250.1 GFDL01006708 JAV28337.1 NEVH01003505 PNF40508.1 JXUM01083118 GAPW01003459 KQ563357 JAC10139.1 KXJ73988.1 AXCM01002898 AAAB01008964 EAA12430.3 APCN01000794 AJVK01022250 AJVK01022251 ATLV01012266 KE524777 KFB36307.1 AJWK01027789 GGFL01000579 MBW64757.1 ADMH02002157 ETN58206.1 GGFJ01008346 MBW57487.1 GGFK01004530 MBW37851.1 GGFK01004554 MBW37875.1 GGFM01003131 MBW23882.1 GL452770 EFN76775.1 GGFJ01008213 MBW57354.1 GL763054 EFZ20220.1 GGFJ01008121 MBW57262.1 KQ971348 EFA05545.1 KQ434977 KZC12941.1 KQ414617 KOC68026.1 GBYB01007560 JAG77327.1 KQ460036 KPJ18422.1 GFDF01010879 JAV03205.1 CH954181 EDV49801.1 KK107126 QOIP01000008 EZA58442.1 RLU19582.1 CM000160 EDW96637.1 GL443590 EFN61854.1 CH477624 EAT38135.1 AE014297 AY051956 AAF54359.1 AAK93380.1 AAN13415.1 AAN13416.1 CH480815 EDW42904.1 CM000364 EDX13645.1 KZ288460 PBC25480.1 KQ776822 OAD52167.1 CM000070 EAL26986.2 OUUW01000005 SPP81163.1 CH902617 EDV42029.1 CH964232 EDW80807.1 CH933806 EDW13955.1 KQ981153 KYN08938.1 CH916378 EDV98546.1 AXCN02000805 GEBQ01012409 GEBQ01004952 JAT27568.1 JAT35025.1 KQ978251 KYM95967.1 CP012526 ALC47131.1 KQ981993 KYN30755.1 CH940650 EDW66512.1 NNAY01002542 OXU21098.1 GEDC01022271 JAS15027.1 KQ982548 KYQ55245.1 KQ976424 KYM88449.1 ADTU01013021 GBXI01002081 GBXI01001770 JAD12211.1 JAD12522.1 GDHF01030024 GDHF01011138 JAI22290.1 JAI41176.1 JXJN01009919 GAKP01009286 GAKP01009285 GAKP01009284 GAKP01009283 GAKP01009282 GAKP01009281 JAC49669.1 JRES01000501 KNC30643.1 CCAG010015282 LJIG01016302 KRT81010.1 GBGD01002659 JAC86230.1 APGK01054668 BT126432 KB741251 KB632404 AEE61396.1 ENN71860.1 ERL95014.1 GFTR01004103 JAW12323.1 GDAI01001136 JAI16467.1 GBBI01002498 JAC16214.1 GGMS01016167 MBY85370.1 GECL01001262 JAP04862.1 GGMR01020312 MBY32931.1 CVRI01000043 CRK95967.1 ABLF02017312 GDHC01015974 JAQ02655.1 GEZM01007120 JAV95366.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000053268
UP000027135
UP000002320
+ More
UP000245037 UP000235965 UP000069940 UP000249989 UP000075884 UP000075883 UP000075885 UP000076408 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000092462 UP000075881 UP000075880 UP000030765 UP000092461 UP000000673 UP000008237 UP000069272 UP000007266 UP000076502 UP000053825 UP000053240 UP000192223 UP000008711 UP000053097 UP000279307 UP000002282 UP000000311 UP000192221 UP000008820 UP000000803 UP000001292 UP000000304 UP000242457 UP000001819 UP000268350 UP000007801 UP000007798 UP000009192 UP000078492 UP000001070 UP000075886 UP000078542 UP000075900 UP000092553 UP000078541 UP000008792 UP000215335 UP000075809 UP000078540 UP000005205 UP000092443 UP000092460 UP000095300 UP000095301 UP000037069 UP000092444 UP000092445 UP000078200 UP000019118 UP000030742 UP000183832 UP000007819
UP000245037 UP000235965 UP000069940 UP000249989 UP000075884 UP000075883 UP000075885 UP000076408 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000092462 UP000075881 UP000075880 UP000030765 UP000092461 UP000000673 UP000008237 UP000069272 UP000007266 UP000076502 UP000053825 UP000053240 UP000192223 UP000008711 UP000053097 UP000279307 UP000002282 UP000000311 UP000192221 UP000008820 UP000000803 UP000001292 UP000000304 UP000242457 UP000001819 UP000268350 UP000007801 UP000007798 UP000009192 UP000078492 UP000001070 UP000075886 UP000078542 UP000075900 UP000092553 UP000078541 UP000008792 UP000215335 UP000075809 UP000078540 UP000005205 UP000092443 UP000092460 UP000095300 UP000095301 UP000037069 UP000092444 UP000092445 UP000078200 UP000019118 UP000030742 UP000183832 UP000007819
Interpro
ProteinModelPortal
A0A2A4JAJ9
A0A2W1BKL3
A0A2H1VY17
A0A212F049
H9JPG1
A0A194PZ27
+ More
U5EPB4 A0A067RBG4 B0WE56 A0A2P8YLX6 A0A1Q3FL93 A0A2J7RI46 A0A023ENW3 A0A182NTT8 A0A182MDE5 A0A182P8K5 A0A182Y3C9 A0A182VIB7 A0A182TII3 A0A182L0S0 A0A182XL97 Q7Q4B4 A0A182ICC7 A0A1B0D1Q8 A0A182K6F0 A0A182ISK4 A0A084VEB3 A0A1B0CTN4 A0A2M4CHD7 W5J306 A0A2M4BWN9 A0A2M4AAN6 A0A2M4AAS5 A0A2M3Z5T7 E2C5G2 A0A2M4BWG9 A0A182F1F2 E9IGP3 A0A2M4BW89 D2A3Q1 A0A154PM36 A0A0L7RBA4 A0A0C9RGT3 A0A0N1IHV2 A0A1L8DA16 A0A1W4WSR6 B3P1R3 A0A026WQX1 B4PUM0 E2AXL6 A0A1W4W1S9 Q16UD3 Q9VHF5 B4HKF4 B4QX74 A0A2A3E1A7 A0A310SCY0 Q29BL1 A0A3B0JK44 B3M072 B4NAJ3 B4K4Y4 A0A195D7U5 B4JYT1 A0A182QZK6 A0A1B6MGJ6 A0A195C523 A0A182RMB3 A0A0M5J7B9 A0A195ER62 B4LW07 A0A232ERV8 A0A1B6CNG7 A0A151X476 A0A195BR62 A0A158NDU4 A0A0A1XMI5 A0A0K8U7J1 A0A1A9Y2K8 A0A1B0B8F0 A0A034W4M5 A0A1I8NR35 A0A1I8MNF0 A0A0L0CE03 A0A1B0FAI5 A0A1A9ZUP4 A0A1A9VGF8 A0A0T6B0Z8 A0A069DQQ4 J3JT76 A0A224XW29 A0A0K8TPZ5 A0A023F567 A0A2S2R6P6 A0A0V0GA21 A0A2S2PTX1 A0A1J1I6N0 J9JKI0 A0A146L4T8 A0A1Y1NGX3
U5EPB4 A0A067RBG4 B0WE56 A0A2P8YLX6 A0A1Q3FL93 A0A2J7RI46 A0A023ENW3 A0A182NTT8 A0A182MDE5 A0A182P8K5 A0A182Y3C9 A0A182VIB7 A0A182TII3 A0A182L0S0 A0A182XL97 Q7Q4B4 A0A182ICC7 A0A1B0D1Q8 A0A182K6F0 A0A182ISK4 A0A084VEB3 A0A1B0CTN4 A0A2M4CHD7 W5J306 A0A2M4BWN9 A0A2M4AAN6 A0A2M4AAS5 A0A2M3Z5T7 E2C5G2 A0A2M4BWG9 A0A182F1F2 E9IGP3 A0A2M4BW89 D2A3Q1 A0A154PM36 A0A0L7RBA4 A0A0C9RGT3 A0A0N1IHV2 A0A1L8DA16 A0A1W4WSR6 B3P1R3 A0A026WQX1 B4PUM0 E2AXL6 A0A1W4W1S9 Q16UD3 Q9VHF5 B4HKF4 B4QX74 A0A2A3E1A7 A0A310SCY0 Q29BL1 A0A3B0JK44 B3M072 B4NAJ3 B4K4Y4 A0A195D7U5 B4JYT1 A0A182QZK6 A0A1B6MGJ6 A0A195C523 A0A182RMB3 A0A0M5J7B9 A0A195ER62 B4LW07 A0A232ERV8 A0A1B6CNG7 A0A151X476 A0A195BR62 A0A158NDU4 A0A0A1XMI5 A0A0K8U7J1 A0A1A9Y2K8 A0A1B0B8F0 A0A034W4M5 A0A1I8NR35 A0A1I8MNF0 A0A0L0CE03 A0A1B0FAI5 A0A1A9ZUP4 A0A1A9VGF8 A0A0T6B0Z8 A0A069DQQ4 J3JT76 A0A224XW29 A0A0K8TPZ5 A0A023F567 A0A2S2R6P6 A0A0V0GA21 A0A2S2PTX1 A0A1J1I6N0 J9JKI0 A0A146L4T8 A0A1Y1NGX3
Ontologies
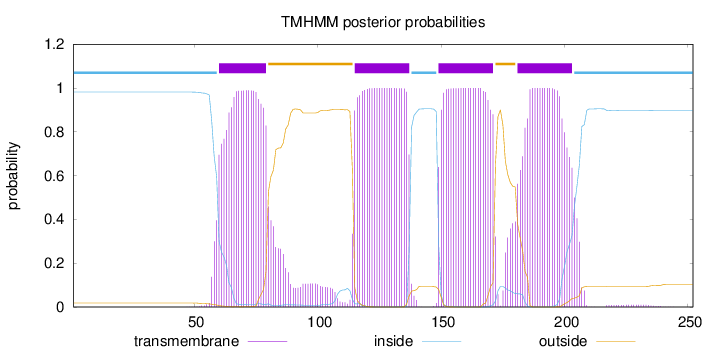
Topology
Length:
252
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
92.82755
Exp number, first 60 AAs:
1.57635
Total prob of N-in:
0.98173
inside
1 - 59
TMhelix
60 - 79
outside
80 - 114
TMhelix
115 - 137
inside
138 - 148
TMhelix
149 - 171
outside
172 - 180
TMhelix
181 - 203
inside
204 - 252
Population Genetic Test Statistics
Pi
29.035401
Theta
17.279542
Tajima's D
-0.542632
CLR
2.462265
CSRT
0.232438378081096
Interpretation
Uncertain
