Pre Gene Modal
BGIBMGA011175
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_diaphanous_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.697
Sequence
CDS
ATGTCCCGACATGAGAAGGCAAAGAGTGGCTTTCTGGACACATGGTTCGGGCGGCCGAAGAAGGCCGGCGCCGGCATCGACCGGGGCTACGACACCGTACCCCGGGGCGCCGAGCCGAGGACGGAAGACGAGGAGCCCAACGCGACCGCGGAGTACACCAGCAAGATAGACGCCCTCGACGACAACCAGCTGCACAGACGATTCGAGGAAATGCTGTCCGACATGAATCTCAACGAGGAGAAAAAGGAGCCGCTCCGGAAATATTCGAGGGATCAGAAAAAGAAGATGCTCGTCGCCTACAAATTCGTTAATACTCAGGAAGGTCGCTCGAAGTTCGAGAAGCCCTCTGATTACGTGGCCTATCTGAACCAGCCCGAGCTGTCTGTCGGGAAACTCCACGGCTGCCTCGAGAACTTGAGGATATCCCTGACTAACAACCCTCTGTCCTGGATCGAAGACTTCGGCACCAAGGGCATCGAGAGTTTATTGACTACCCTCAACTTGTGTTACACGAACGATTCCCGTTACGACAGAGTCCAGTACGAGTGCATAAGATGTTTGTCGGCGATCCTGAACAACACGGTCGGTATTAGGACGATGTTCGACTGCAGGGAGGCCCTGCCGGTCTTGGCGAGGAGTCTGGACGCCAGGAAGCCACATTGCGCACTAGAGGCCGCCAAGGTGTTAGCGGCCATATGTCTGATACCGAACGGGCACGAGAAGGTGCTGGAAGCCATAACGATGGCCGGCGAGTCCAGCAGGAAAGCTAGGTTCCTGCCCATCGTCGAAGGACTGGAAAGCAAAAGCCCGGAGAGCTTGAAGAACGGCTGCATGCAATTGATGAACGCCATCATAACGGAGCCCGAAGAGTTGGAGTTCAGGCTGCATCTGAGGAGCGAGTTCATGAGGACGGGCCTCTACGACCTCATAGACGAACTGCGCGCCGGCGCTGAAGGCTCCCGCCTCATACAAGTGAACGTGTTCGACTCGCACGCGTCCGCCGACCAGGACGAATTCCTGGCGAAATTCGACGACGTCAGGGTCGACTTCAACGATTCGAACGAGTGCTTCGAGCTGGTGAAGAACCTGGTCATAGATACGCCGGCGGAGCCCTACCTGCTGTCCATACTGCAGCACTTGCTGTTCATAAGAGATGACGAGTTGATCAGGCCGGCCTACTACAAGCTGATTGAGGAGTGCATCACGCAGATAGTCCTGCACAAGAACGGCTACGACCCAGACTTCAGGGCGACGCAGCGGTTCAACATAGACGTGCAGCCGCTCATCGACGGACTCATAGAAAAATCTCGAGCCGAAGAAGAGAAGAGGTTGGAGGAGTTGAAGTCGAAGCTGGAGGCGGCGATCGCGGCCAGACAGGAGGCCGAGGCGAGGGCCGCCCACCTGGAGGAGAGGCTGAAGAGTACTGCCTCCCCCACCGCGCCCGGCGGCCTCTCGCAAGGGAACATCGCCGCCATTGCCAAGGTACACGCGACCACTGGTCAAAACAAACCTAACCAGTCCAAAGCTGTTGGGTACGTGTCACACGTGTCGATAAGGCGAGTTTCGTCATCCCAGGTTGGACGGGTTTATTAA
Protein
MSRHEKAKSGFLDTWFGRPKKAGAGIDRGYDTVPRGAEPRTEDEEPNATAEYTSKIDALDDNQLHRRFEEMLSDMNLNEEKKEPLRKYSRDQKKKMLVAYKFVNTQEGRSKFEKPSDYVAYLNQPELSVGKLHGCLENLRISLTNNPLSWIEDFGTKGIESLLTTLNLCYTNDSRYDRVQYECIRCLSAILNNTVGIRTMFDCREALPVLARSLDARKPHCALEAAKVLAAICLIPNGHEKVLEAITMAGESSRKARFLPIVEGLESKSPESLKNGCMQLMNAIITEPEELEFRLHLRSEFMRTGLYDLIDELRAGAEGSRLIQVNVFDSHASADQDEFLAKFDDVRVDFNDSNECFELVKNLVIDTPAEPYLLSILQHLLFIRDDELIRPAYYKLIEECITQIVLHKNGYDPDFRATQRFNIDVQPLIDGLIEKSRAEEEKRLEELKSKLEAAIAARQEAEARAAHLEERLKSTASPTAPGGLSQGNIAAIAKVHATTGQNKPNQSKAVGYVSHVSIRRVSSSQVGRVY
Summary
Uniprot
A0A194PSX1
H9JNS1
A0A212F036
A0A2W1BSF6
A0A2J7RNA0
A0A2J7RNA2
+ More
F4WKK9 A0A2J7RNA5 K7J3T0 A0A0C9R6Z4 A0A0C9QXL0 A0A026X427 E2AKP6 A0A151JBQ0 A0A2S2PNK7 E0VPZ1 A0A088AU80 A0A151JW76 A0A2A3EPN3 A0A3L8E0T9 A0A2P8XDR6 A0A195C2V4 A0A3Q0J2S6 E2BHB5 A0A0A9XQV0 A0A0L7RB02 A0A2H8TDF6 A0A2S2QCC5 A0A158NNE7 J9JPD5 A0A0J7LA64 A0A146LN18 A0A146LK79 A0A0A9XT57 A0A232FLG2 A0A2J7RNB5 A0A0K2UZ14 A0A0M9A4C3 A0A0P5S4L9 A0A0P5RYR2 A0A0P5SK41 A0A0P6GBK6 A0A0P5YYL8 A0A0P5SFH2 A0A0P5K4Y6 E9FT11 A0A0N8C6K5 A0A0P5LM82 A0A0N8AQT5 A0A0P6G7M6 A0A0P5FZA4 A0A0P5HZ33 A0A0P4ZJF1 A0A0P4ZK55 A0A0P5H8M9 A0A0P6E706 A0A0P4YI36 A0A0N8BBY3 A0A0P5TAC4 A0A0P5W8D9 A0A0P5SRT7 A0A0P6AN41 A0A0P5THE0 A0A0P5KBW3 A0A0N8AHC0 A0A0P5G8W5 A0A0P6I8A7 A0A0P4ZKA9 A0A0P4ZTV9 A0A0N8CBZ8 A0A0N8CW78 A0A0P5TR44 A0A0P6GRC6 A0A0P5E8A8 A0A0P5SCZ7 A0A0P6B109 A0A0P4XXK7 A0A0P5TWB5 A0A162DCJ0 A0A1Y1MKZ6 A0A0P5XD44 T1IBQ7 A0A2S2Q532 A0A1Y1MQX8 A0A1Y1MPJ2 A0A1Y1MKY4 A0A1S3IN32 A0A2R5L4H2 A0A1Z5LFA7 A0A210QAN3 A0A1D8KD26 A0A1D2MVR6
F4WKK9 A0A2J7RNA5 K7J3T0 A0A0C9R6Z4 A0A0C9QXL0 A0A026X427 E2AKP6 A0A151JBQ0 A0A2S2PNK7 E0VPZ1 A0A088AU80 A0A151JW76 A0A2A3EPN3 A0A3L8E0T9 A0A2P8XDR6 A0A195C2V4 A0A3Q0J2S6 E2BHB5 A0A0A9XQV0 A0A0L7RB02 A0A2H8TDF6 A0A2S2QCC5 A0A158NNE7 J9JPD5 A0A0J7LA64 A0A146LN18 A0A146LK79 A0A0A9XT57 A0A232FLG2 A0A2J7RNB5 A0A0K2UZ14 A0A0M9A4C3 A0A0P5S4L9 A0A0P5RYR2 A0A0P5SK41 A0A0P6GBK6 A0A0P5YYL8 A0A0P5SFH2 A0A0P5K4Y6 E9FT11 A0A0N8C6K5 A0A0P5LM82 A0A0N8AQT5 A0A0P6G7M6 A0A0P5FZA4 A0A0P5HZ33 A0A0P4ZJF1 A0A0P4ZK55 A0A0P5H8M9 A0A0P6E706 A0A0P4YI36 A0A0N8BBY3 A0A0P5TAC4 A0A0P5W8D9 A0A0P5SRT7 A0A0P6AN41 A0A0P5THE0 A0A0P5KBW3 A0A0N8AHC0 A0A0P5G8W5 A0A0P6I8A7 A0A0P4ZKA9 A0A0P4ZTV9 A0A0N8CBZ8 A0A0N8CW78 A0A0P5TR44 A0A0P6GRC6 A0A0P5E8A8 A0A0P5SCZ7 A0A0P6B109 A0A0P4XXK7 A0A0P5TWB5 A0A162DCJ0 A0A1Y1MKZ6 A0A0P5XD44 T1IBQ7 A0A2S2Q532 A0A1Y1MQX8 A0A1Y1MPJ2 A0A1Y1MKY4 A0A1S3IN32 A0A2R5L4H2 A0A1Z5LFA7 A0A210QAN3 A0A1D8KD26 A0A1D2MVR6
Pubmed
EMBL
KQ459593
KPI96412.1
BABH01010022
BABH01010023
AGBW02011174
OWR47100.1
+ More
KZ149991 PZC75576.1 NEVH01002545 PNF42313.1 PNF42314.1 GL888202 EGI65259.1 PNF42316.1 GBYB01003715 JAG73482.1 GBYB01008519 JAG78286.1 KK107015 EZA62858.1 GL440368 EFN65986.1 KQ979171 KYN22375.1 GGMR01018432 MBY31051.1 DS235379 EEB15447.1 KQ981661 KYN38521.1 KZ288203 PBC33464.1 QOIP01000002 RLU25638.1 PYGN01002652 PSN30139.1 KQ978379 KYM94518.1 GL448287 EFN84884.1 GBHO01038821 GBHO01021598 GBHO01021596 GBHO01021593 GBRD01015991 JAG04783.1 JAG22006.1 JAG22008.1 JAG22011.1 JAG49835.1 KQ414617 KOC68044.1 GFXV01000314 MBW12119.1 GGMS01006160 MBY75363.1 ADTU01002697 ADTU01002698 ABLF02031314 LBMM01000070 KMR05081.1 GDHC01009298 JAQ09331.1 GDHC01010251 GDHC01009212 JAQ08378.1 JAQ09417.1 GBHO01038826 GBHO01021600 GBHO01021599 GBHO01021591 JAG04778.1 JAG22004.1 JAG22005.1 JAG22013.1 NNAY01000066 OXU31350.1 PNF42312.1 HACA01025959 CDW43320.1 KQ435752 KOX76206.1 GDIQ01092474 JAL59252.1 GDIQ01094674 JAL57052.1 GDIQ01094673 JAL57053.1 GDIQ01046047 JAN48690.1 GDIP01051388 JAM52327.1 GDIQ01092473 JAL59253.1 GDIQ01190648 JAK61077.1 GL732524 EFX89725.1 GDIQ01109819 JAL41907.1 GDIQ01171544 JAK80181.1 GDIQ01251979 JAJ99745.1 GDIQ01038904 JAN55833.1 GDIQ01249034 JAK02691.1 GDIQ01221533 JAK30192.1 GDIP01212463 JAJ10939.1 GDIP01211565 JAJ11837.1 GDIQ01233532 JAK18193.1 GDIQ01078677 JAN16060.1 GDIP01229669 JAI93732.1 GDIQ01192795 JAK58930.1 GDIQ01094672 JAL57054.1 GDIP01089781 JAM13934.1 GDIP01141474 JAL62240.1 GDIP01040585 JAM63130.1 GDIP01132078 JAL71636.1 GDIQ01192794 JAK58931.1 GDIP01147377 JAJ76025.1 GDIQ01251980 JAJ99744.1 GDIQ01008111 JAN86626.1 GDIP01212464 JAJ10938.1 GDIP01211566 JAJ11836.1 GDIQ01094675 JAL57051.1 GDIP01092823 JAM10892.1 GDIP01122828 GDIP01064028 JAL80886.1 GDIQ01184744 GDIQ01029711 JAN65026.1 GDIP01163516 JAJ59886.1 GDIP01141475 JAL62239.1 GDIP01021656 JAM82059.1 GDIP01235364 JAI88037.1 GDIP01121075 JAL82639.1 LRGB01001937 KZS09914.1 GEZM01028215 JAV86374.1 GDIP01073889 JAM29826.1 ACPB03004993 GGMS01003660 MBY72863.1 GEZM01028213 JAV86376.1 GEZM01028218 GEZM01028214 JAV86375.1 GEZM01028216 JAV86373.1 GGLE01000270 MBY04396.1 GFJQ02001359 JAW05611.1 NEDP02004404 OWF45792.1 KX387874 AOV18876.1 LJIJ01000465 ODM97180.1
KZ149991 PZC75576.1 NEVH01002545 PNF42313.1 PNF42314.1 GL888202 EGI65259.1 PNF42316.1 GBYB01003715 JAG73482.1 GBYB01008519 JAG78286.1 KK107015 EZA62858.1 GL440368 EFN65986.1 KQ979171 KYN22375.1 GGMR01018432 MBY31051.1 DS235379 EEB15447.1 KQ981661 KYN38521.1 KZ288203 PBC33464.1 QOIP01000002 RLU25638.1 PYGN01002652 PSN30139.1 KQ978379 KYM94518.1 GL448287 EFN84884.1 GBHO01038821 GBHO01021598 GBHO01021596 GBHO01021593 GBRD01015991 JAG04783.1 JAG22006.1 JAG22008.1 JAG22011.1 JAG49835.1 KQ414617 KOC68044.1 GFXV01000314 MBW12119.1 GGMS01006160 MBY75363.1 ADTU01002697 ADTU01002698 ABLF02031314 LBMM01000070 KMR05081.1 GDHC01009298 JAQ09331.1 GDHC01010251 GDHC01009212 JAQ08378.1 JAQ09417.1 GBHO01038826 GBHO01021600 GBHO01021599 GBHO01021591 JAG04778.1 JAG22004.1 JAG22005.1 JAG22013.1 NNAY01000066 OXU31350.1 PNF42312.1 HACA01025959 CDW43320.1 KQ435752 KOX76206.1 GDIQ01092474 JAL59252.1 GDIQ01094674 JAL57052.1 GDIQ01094673 JAL57053.1 GDIQ01046047 JAN48690.1 GDIP01051388 JAM52327.1 GDIQ01092473 JAL59253.1 GDIQ01190648 JAK61077.1 GL732524 EFX89725.1 GDIQ01109819 JAL41907.1 GDIQ01171544 JAK80181.1 GDIQ01251979 JAJ99745.1 GDIQ01038904 JAN55833.1 GDIQ01249034 JAK02691.1 GDIQ01221533 JAK30192.1 GDIP01212463 JAJ10939.1 GDIP01211565 JAJ11837.1 GDIQ01233532 JAK18193.1 GDIQ01078677 JAN16060.1 GDIP01229669 JAI93732.1 GDIQ01192795 JAK58930.1 GDIQ01094672 JAL57054.1 GDIP01089781 JAM13934.1 GDIP01141474 JAL62240.1 GDIP01040585 JAM63130.1 GDIP01132078 JAL71636.1 GDIQ01192794 JAK58931.1 GDIP01147377 JAJ76025.1 GDIQ01251980 JAJ99744.1 GDIQ01008111 JAN86626.1 GDIP01212464 JAJ10938.1 GDIP01211566 JAJ11836.1 GDIQ01094675 JAL57051.1 GDIP01092823 JAM10892.1 GDIP01122828 GDIP01064028 JAL80886.1 GDIQ01184744 GDIQ01029711 JAN65026.1 GDIP01163516 JAJ59886.1 GDIP01141475 JAL62239.1 GDIP01021656 JAM82059.1 GDIP01235364 JAI88037.1 GDIP01121075 JAL82639.1 LRGB01001937 KZS09914.1 GEZM01028215 JAV86374.1 GDIP01073889 JAM29826.1 ACPB03004993 GGMS01003660 MBY72863.1 GEZM01028213 JAV86376.1 GEZM01028218 GEZM01028214 JAV86375.1 GEZM01028216 JAV86373.1 GGLE01000270 MBY04396.1 GFJQ02001359 JAW05611.1 NEDP02004404 OWF45792.1 KX387874 AOV18876.1 LJIJ01000465 ODM97180.1
Proteomes
UP000053268
UP000005204
UP000007151
UP000235965
UP000007755
UP000002358
+ More
UP000053097 UP000000311 UP000078492 UP000009046 UP000005203 UP000078541 UP000242457 UP000279307 UP000245037 UP000078542 UP000079169 UP000008237 UP000053825 UP000005205 UP000007819 UP000036403 UP000215335 UP000053105 UP000000305 UP000076858 UP000015103 UP000085678 UP000242188 UP000094527
UP000053097 UP000000311 UP000078492 UP000009046 UP000005203 UP000078541 UP000242457 UP000279307 UP000245037 UP000078542 UP000079169 UP000008237 UP000053825 UP000005205 UP000007819 UP000036403 UP000215335 UP000053105 UP000000305 UP000076858 UP000015103 UP000085678 UP000242188 UP000094527
PRIDE
Pfam
Interpro
IPR042201
FH2_Formin_sf
+ More
IPR010472 FH3_dom
IPR015425 FH2_Formin
IPR014767 DAD_dom
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR010473 GTPase-bd
IPR014768 GBD/FH3_dom
IPR010465 Drf_DAD
IPR027654 Formin_DIAPH3
IPR027644 DIAPH2
IPR011072 HR1_rho-bd
IPR001478 PDZ
IPR036034 PDZ_sf
IPR036274 HR1_rpt_sf
IPR038499 BRO1_sf
IPR004328 BRO1_dom
IPR027653 Formin_Diaph1
IPR009408 Formin_homology_1
IPR010472 FH3_dom
IPR015425 FH2_Formin
IPR014767 DAD_dom
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR010473 GTPase-bd
IPR014768 GBD/FH3_dom
IPR010465 Drf_DAD
IPR027654 Formin_DIAPH3
IPR027644 DIAPH2
IPR011072 HR1_rho-bd
IPR001478 PDZ
IPR036034 PDZ_sf
IPR036274 HR1_rpt_sf
IPR038499 BRO1_sf
IPR004328 BRO1_dom
IPR027653 Formin_Diaph1
IPR009408 Formin_homology_1
Gene 3D
ProteinModelPortal
A0A194PSX1
H9JNS1
A0A212F036
A0A2W1BSF6
A0A2J7RNA0
A0A2J7RNA2
+ More
F4WKK9 A0A2J7RNA5 K7J3T0 A0A0C9R6Z4 A0A0C9QXL0 A0A026X427 E2AKP6 A0A151JBQ0 A0A2S2PNK7 E0VPZ1 A0A088AU80 A0A151JW76 A0A2A3EPN3 A0A3L8E0T9 A0A2P8XDR6 A0A195C2V4 A0A3Q0J2S6 E2BHB5 A0A0A9XQV0 A0A0L7RB02 A0A2H8TDF6 A0A2S2QCC5 A0A158NNE7 J9JPD5 A0A0J7LA64 A0A146LN18 A0A146LK79 A0A0A9XT57 A0A232FLG2 A0A2J7RNB5 A0A0K2UZ14 A0A0M9A4C3 A0A0P5S4L9 A0A0P5RYR2 A0A0P5SK41 A0A0P6GBK6 A0A0P5YYL8 A0A0P5SFH2 A0A0P5K4Y6 E9FT11 A0A0N8C6K5 A0A0P5LM82 A0A0N8AQT5 A0A0P6G7M6 A0A0P5FZA4 A0A0P5HZ33 A0A0P4ZJF1 A0A0P4ZK55 A0A0P5H8M9 A0A0P6E706 A0A0P4YI36 A0A0N8BBY3 A0A0P5TAC4 A0A0P5W8D9 A0A0P5SRT7 A0A0P6AN41 A0A0P5THE0 A0A0P5KBW3 A0A0N8AHC0 A0A0P5G8W5 A0A0P6I8A7 A0A0P4ZKA9 A0A0P4ZTV9 A0A0N8CBZ8 A0A0N8CW78 A0A0P5TR44 A0A0P6GRC6 A0A0P5E8A8 A0A0P5SCZ7 A0A0P6B109 A0A0P4XXK7 A0A0P5TWB5 A0A162DCJ0 A0A1Y1MKZ6 A0A0P5XD44 T1IBQ7 A0A2S2Q532 A0A1Y1MQX8 A0A1Y1MPJ2 A0A1Y1MKY4 A0A1S3IN32 A0A2R5L4H2 A0A1Z5LFA7 A0A210QAN3 A0A1D8KD26 A0A1D2MVR6
F4WKK9 A0A2J7RNA5 K7J3T0 A0A0C9R6Z4 A0A0C9QXL0 A0A026X427 E2AKP6 A0A151JBQ0 A0A2S2PNK7 E0VPZ1 A0A088AU80 A0A151JW76 A0A2A3EPN3 A0A3L8E0T9 A0A2P8XDR6 A0A195C2V4 A0A3Q0J2S6 E2BHB5 A0A0A9XQV0 A0A0L7RB02 A0A2H8TDF6 A0A2S2QCC5 A0A158NNE7 J9JPD5 A0A0J7LA64 A0A146LN18 A0A146LK79 A0A0A9XT57 A0A232FLG2 A0A2J7RNB5 A0A0K2UZ14 A0A0M9A4C3 A0A0P5S4L9 A0A0P5RYR2 A0A0P5SK41 A0A0P6GBK6 A0A0P5YYL8 A0A0P5SFH2 A0A0P5K4Y6 E9FT11 A0A0N8C6K5 A0A0P5LM82 A0A0N8AQT5 A0A0P6G7M6 A0A0P5FZA4 A0A0P5HZ33 A0A0P4ZJF1 A0A0P4ZK55 A0A0P5H8M9 A0A0P6E706 A0A0P4YI36 A0A0N8BBY3 A0A0P5TAC4 A0A0P5W8D9 A0A0P5SRT7 A0A0P6AN41 A0A0P5THE0 A0A0P5KBW3 A0A0N8AHC0 A0A0P5G8W5 A0A0P6I8A7 A0A0P4ZKA9 A0A0P4ZTV9 A0A0N8CBZ8 A0A0N8CW78 A0A0P5TR44 A0A0P6GRC6 A0A0P5E8A8 A0A0P5SCZ7 A0A0P6B109 A0A0P4XXK7 A0A0P5TWB5 A0A162DCJ0 A0A1Y1MKZ6 A0A0P5XD44 T1IBQ7 A0A2S2Q532 A0A1Y1MQX8 A0A1Y1MPJ2 A0A1Y1MKY4 A0A1S3IN32 A0A2R5L4H2 A0A1Z5LFA7 A0A210QAN3 A0A1D8KD26 A0A1D2MVR6
PDB
1Z2C
E-value=5.93342e-82,
Score=776
Ontologies
GO
PANTHER
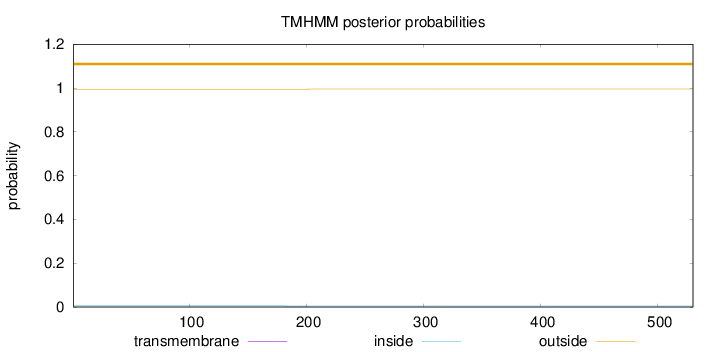
Topology
Length:
530
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00802999999999997
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00523
outside
1 - 530
Population Genetic Test Statistics
Pi
20.447182
Theta
19.161105
Tajima's D
0.229248
CLR
0.593063
CSRT
0.44012799360032
Interpretation
Uncertain
