Gene
KWMTBOMO14161
Pre Gene Modal
BGIBMGA011177
Annotation
PREDICTED:_43_kDa_receptor-associated_protein_of_the_synapse_homolog?_partial_[Bombyx_mori]
Full name
P-type phospholipid transporter
+ More
43 kDa receptor-associated protein of the synapse homolog
43 kDa receptor-associated protein of the synapse homolog
Location in the cell
Mitochondrial Reliability : 1.851 Nuclear Reliability : 1.415
Sequence
CDS
ATGTCGTGGGACTCTATCGAGTCACGTGACTTTCTCGGCGGAGTGCCGGCGCATCAGTTATCAGCCACACGTCTTCTCAGTTCACCAGATCCGTCGAGACACCATCTCGATGAGACGCAAGAAGTATATCCATTTTCGGAGGAATACCCAGGACCGGCTTCGGCCAGGAATGGCGCGTTGCGCTGGGGCGCGAGGTTCGGTCCGGCCCACCGACCACCGGCCTCTGGGCTGCTGGATTGCTTCCGAGTTTGCCGGTCTAGACTCGACCAGTACCTGGCCAGGCGGAAGATTGAACGCGGGGTCCGTCTGTACGGTCAGAACGAGCAGCGCCAGGCCGTCAAGGTATGGCTGTCAGCGTTGCGCGGCGTCAGACACAGAAACGACAAGTTCGCCCTCTTAGGACATCTGTACCAGGCGTACATGGACTTTGGGAAGTACAGAGAATCGTTGGAGTTCGCAAACAGACAACTAGGTATTTCCGAAGAGCTGGACTCCGCTCCGATGCGGGCCGAGGCGTACCTCAACTTGGCGAGGGCTCACCAAACCCTGGGCGGGCTGGACAGAGCTCTCTCGTACGCCCGCCACGCTCTGTACAACGACTGCGGCGGCGGCTCCACGGCCGGCTACGTCCACCTCACCGTGGCCTCCGTGAGCTTAGAGCTCGGAGCTTTCTCCAAGGCCATGGACGGCTTCCAGAAAGCTCTTGCAGTGGCGCAGGCGCAGAACGACAACACGCTATTGCTACAGGTGTACGTGGGTCTGTCTGAGCTGTGGCGGCGGCTGCGGGACGCGGAGCGCGCGGTGGCGTGCGCGGCGCGGGCCTGCGACCTGGGCCGGGGCCCGCGCGCCGCCGACCTCAACACGCGTCACCACCGCACGGCGCTGCTGCAAATGGCGGCCGCCTTGCGCGCCCGCGGCGAGCTCGGAGACGCGCACGACTACTGCAACGAGGCGCTGCGGCTGGCGGCGGTGGCCGGGGACCAGGGCTGCTACGCTCGCGCCGTCCGCATCGCCGGCGACGTCTACCGACGGAAGTGCGACGTCGGCAAAGCTCTCAGGCACTACGAAGTGGCCATGGGCGCGGGCCAGGCATTAGGCGACCGTCTGGCCCAGATGGAGGCCATGGACGGCGCCGCGCGCTGCCTGGAGGCTCTCCGTCTGCAGAACAGGATCTGCAACTGTCGTCCGCTCGAGTTCAACACGAGATTACTGGAAGTCGCCACTTCCGTCGGAGCCAAGATGCTGGTACGACGGGTTCGACTCCGCCTAGCCGAGATCTACACCGCGTTGGGCGACAACAAGGCTGCAGCGAGGCAGAAAAGAGCCGCGGCCGCCTGGGGAGCCCCGGCCTGCGCCCGATGCAGGGAGCCGCTCGCGGAGCAGCCCGAGCCGCTGGCGGCGTTGCCGTGCGCGCACATCGTGCACCACGCGTGA
Protein
MSWDSIESRDFLGGVPAHQLSATRLLSSPDPSRHHLDETQEVYPFSEEYPGPASARNGALRWGARFGPAHRPPASGLLDCFRVCRSRLDQYLARRKIERGVRLYGQNEQRQAVKVWLSALRGVRHRNDKFALLGHLYQAYMDFGKYRESLEFANRQLGISEELDSAPMRAEAYLNLARAHQTLGGLDRALSYARHALYNDCGGGSTAGYVHLTVASVSLELGAFSKAMDGFQKALAVAQAQNDNTLLLQVYVGLSELWRRLRDAERAVACAARACDLGRGPRAADLNTRHHRTALLQMAAALRARGELGDAHDYCNEALRLAAVAGDQGCYARAVRIAGDVYRRKCDVGKALRHYEVAMGAGQALGDRLAQMEAMDGAARCLEALRLQNRICNCRPLEFNTRLLEVATSVGAKMLVRRVRLRLAEIYTALGDNKAAARQKRAAAAWGAPACARCREPLAEQPEPLAALPCAHIVHHA
Summary
Description
Postsynaptic protein required for clustering of nicotinic acetylcholine receptors (nAChRs) at the neuromuscular junction.
Catalytic Activity
ATP + H(2)O + phospholipid(Side 1) = ADP + phosphate + phospholipid(Side 2).
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IV subfamily.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the RAPsyn family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the RAPsyn family.
Keywords
Complete proteome
Metal-binding
Reference proteome
Repeat
TPR repeat
Ubl conjugation
Zinc
Zinc-finger
Feature
chain 43 kDa receptor-associated protein of the synapse homolog
Uniprot
A0A0N0PDY7
H9JNS3
A0A2H1VFP7
A0A0C9S063
D2A4B4
A0A088AU77
+ More
A0A2A3EP21 A0A3L8DIN3 W8BUF7 W8CDP2 A0A034WI23 A0A0J9S128 B4NHM1 B4IIX3 B3P9S6 F4WQN1 Q7K4Y8 B4PW15 A0A151WV73 E2A7I6 E2BYH2 A0A3B0KT43 A0A1W4VIM2 B4MEZ9 A0A3B0K4F4 A0A310SFW0 B4H875 Q29CY0 K7IML0 B4JZW5 B4L7B4 A0A026WPI6 A0A195D194 A0A0L7RAU4 A0A0J9S0T6 A0A195BRF3 B3MZZ4 A0A2J7PLY3 A0A0M4ES75 A0A158NRZ9 A0A195ERG6 E9IHY2 A0A0M9A2Y4 A0A195D819 A0A1J1HXJ0 A0A1J1HZI4 E0VSU8 A0A336MFR4 A0A3B0K482 A0A1W4VHV8 A0A0A9XSG1 A0A2P8Y3Y9 B4R2F8 U4UZJ4 T1J5G7 R7UF25 A0A0N5ATC2 A0A2A6CT05 A0A0N4V4A9 A0A2G9V5Q0 A0A016UEW2 F1KZS1 A0A1I7TV07 A0A0N4UKH6 A0A2A2LPU3 E3LQX7 A8XCH4 G0NR17 A0A2A2LPD5 A0A2P4V7M8 A0A261BPL8 A0A1S3H1K5 A0A2A2LPB4 Q09485 A0A2A2LP52 A0A0B2V6M7 A0A183V9A1 A0A2A2JR22 K7H5E4 A0A1I7VLE5 A0A0N4YCI1 A0A2G5UZP1 A0A158PFP0 A0A2G5UZP6 A0A1I7RPI5 A0A0N4TVD9 A0A2G5UZR7 A0A044VIV9
A0A2A3EP21 A0A3L8DIN3 W8BUF7 W8CDP2 A0A034WI23 A0A0J9S128 B4NHM1 B4IIX3 B3P9S6 F4WQN1 Q7K4Y8 B4PW15 A0A151WV73 E2A7I6 E2BYH2 A0A3B0KT43 A0A1W4VIM2 B4MEZ9 A0A3B0K4F4 A0A310SFW0 B4H875 Q29CY0 K7IML0 B4JZW5 B4L7B4 A0A026WPI6 A0A195D194 A0A0L7RAU4 A0A0J9S0T6 A0A195BRF3 B3MZZ4 A0A2J7PLY3 A0A0M4ES75 A0A158NRZ9 A0A195ERG6 E9IHY2 A0A0M9A2Y4 A0A195D819 A0A1J1HXJ0 A0A1J1HZI4 E0VSU8 A0A336MFR4 A0A3B0K482 A0A1W4VHV8 A0A0A9XSG1 A0A2P8Y3Y9 B4R2F8 U4UZJ4 T1J5G7 R7UF25 A0A0N5ATC2 A0A2A6CT05 A0A0N4V4A9 A0A2G9V5Q0 A0A016UEW2 F1KZS1 A0A1I7TV07 A0A0N4UKH6 A0A2A2LPU3 E3LQX7 A8XCH4 G0NR17 A0A2A2LPD5 A0A2P4V7M8 A0A261BPL8 A0A1S3H1K5 A0A2A2LPB4 Q09485 A0A2A2LP52 A0A0B2V6M7 A0A183V9A1 A0A2A2JR22 K7H5E4 A0A1I7VLE5 A0A0N4YCI1 A0A2G5UZP1 A0A158PFP0 A0A2G5UZP6 A0A1I7RPI5 A0A0N4TVD9 A0A2G5UZR7 A0A044VIV9
EC Number
7.6.2.1
Pubmed
26354079
19121390
18362917
19820115
30249741
24495485
+ More
25348373 22936249 17994087 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20798317 18057021 20479145 15632085 23185243 20075255 24508170 21347285 21282665 20566863 25401762 26823975 29403074 23537049 23254933 18806794 25730766 21685128 14624247 26114425 26394399 9851916 19158078 25217238 21909270
25348373 22936249 17994087 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20798317 18057021 20479145 15632085 23185243 20075255 24508170 21347285 21282665 20566863 25401762 26823975 29403074 23537049 23254933 18806794 25730766 21685128 14624247 26114425 26394399 9851916 19158078 25217238 21909270
EMBL
KQ460036
KPJ18415.1
BABH01010020
BABH01010021
ODYU01002323
SOQ39668.1
+ More
GBYB01013846 JAG83613.1 KQ971348 EFA04813.2 KZ288203 PBC33467.1 QOIP01000007 RLU20294.1 GAMC01001600 JAC04956.1 GAMC01001602 GAMC01001601 GAMC01001599 JAC04955.1 GAKP01005167 GAKP01005166 JAC53785.1 CM002916 KMZ07253.1 CH964272 EDW83590.2 CH480845 EDW50914.1 CH954184 EDV45239.1 GL888275 EGI63394.1 AE014135 AY051576 AAF59332.2 AAF59333.2 AAK93000.1 CM000161 EDW99320.1 KRK04908.1 KQ982711 KYQ51754.1 GL437329 EFN70683.1 GL451478 EFN79256.1 OUUW01000017 SPP89026.1 CM000831 CH940665 ACY70527.1 EDW71100.1 KRF85349.1 SPP89025.1 KQ775154 OAD52290.1 CH479221 EDW34875.1 CH475397 EAL29280.1 KRT05198.1 KRT05199.1 CH916380 EDV90981.1 CH933813 EDW10908.1 KRG07266.1 KK107152 EZA57004.1 KQ977041 KYN06174.1 KQ414617 KOC68042.1 KMZ07252.1 KQ976424 KYM88536.1 CH902637 EDV44785.2 NEVH01024421 PNF17345.1 CP012527 ALC48126.1 ADTU01024378 ADTU01024379 KQ981993 KYN30850.1 GL763300 EFZ19824.1 KQ435752 KOX76205.1 KQ981153 KYN09035.1 CVRI01000035 CRK92741.1 CRK92740.1 DS235758 EEB16454.1 UFQT01001076 SSX28770.1 SPP89027.1 GBHO01020705 GDHC01018505 GDHC01010813 JAG22899.1 JAQ00124.1 JAQ07816.1 PYGN01000955 PSN38963.1 CM000365 EDX15209.1 KI208086 ERL95790.1 JH431861 AMQN01001390 KB302197 ELU04574.1 ABKE03000020 PDM81354.1 UXUI01007913 VDD89887.1 KZ344986 PIO77799.1 JARK01001381 EYC13138.1 JI168699 ADY43375.1 UYYG01000053 VDN52321.1 LIAE01006535 PAV87997.1 DS268413 NMWX01000827 EFP07774.1 OZF76061.1 HE600914 CAP30416.2 GL379929 EGT36014.1 PAV87999.1 LFJK02001230 POM33298.1 NIPN01000105 OZG11665.1 PAV87998.1 FO080625 PAV87996.1 JPKZ01002363 KHN77154.1 UYWY01024328 VDM48642.1 LIAE01010286 PAV64061.1 UYSL01021291 VDL77839.1 PDUG01000002 PIC44997.1 UYYA01003809 VDM55829.1 PIC44998.1 UZAD01013320 VDN93953.1 PIC44999.1 CMVM020000306
GBYB01013846 JAG83613.1 KQ971348 EFA04813.2 KZ288203 PBC33467.1 QOIP01000007 RLU20294.1 GAMC01001600 JAC04956.1 GAMC01001602 GAMC01001601 GAMC01001599 JAC04955.1 GAKP01005167 GAKP01005166 JAC53785.1 CM002916 KMZ07253.1 CH964272 EDW83590.2 CH480845 EDW50914.1 CH954184 EDV45239.1 GL888275 EGI63394.1 AE014135 AY051576 AAF59332.2 AAF59333.2 AAK93000.1 CM000161 EDW99320.1 KRK04908.1 KQ982711 KYQ51754.1 GL437329 EFN70683.1 GL451478 EFN79256.1 OUUW01000017 SPP89026.1 CM000831 CH940665 ACY70527.1 EDW71100.1 KRF85349.1 SPP89025.1 KQ775154 OAD52290.1 CH479221 EDW34875.1 CH475397 EAL29280.1 KRT05198.1 KRT05199.1 CH916380 EDV90981.1 CH933813 EDW10908.1 KRG07266.1 KK107152 EZA57004.1 KQ977041 KYN06174.1 KQ414617 KOC68042.1 KMZ07252.1 KQ976424 KYM88536.1 CH902637 EDV44785.2 NEVH01024421 PNF17345.1 CP012527 ALC48126.1 ADTU01024378 ADTU01024379 KQ981993 KYN30850.1 GL763300 EFZ19824.1 KQ435752 KOX76205.1 KQ981153 KYN09035.1 CVRI01000035 CRK92741.1 CRK92740.1 DS235758 EEB16454.1 UFQT01001076 SSX28770.1 SPP89027.1 GBHO01020705 GDHC01018505 GDHC01010813 JAG22899.1 JAQ00124.1 JAQ07816.1 PYGN01000955 PSN38963.1 CM000365 EDX15209.1 KI208086 ERL95790.1 JH431861 AMQN01001390 KB302197 ELU04574.1 ABKE03000020 PDM81354.1 UXUI01007913 VDD89887.1 KZ344986 PIO77799.1 JARK01001381 EYC13138.1 JI168699 ADY43375.1 UYYG01000053 VDN52321.1 LIAE01006535 PAV87997.1 DS268413 NMWX01000827 EFP07774.1 OZF76061.1 HE600914 CAP30416.2 GL379929 EGT36014.1 PAV87999.1 LFJK02001230 POM33298.1 NIPN01000105 OZG11665.1 PAV87998.1 FO080625 PAV87996.1 JPKZ01002363 KHN77154.1 UYWY01024328 VDM48642.1 LIAE01010286 PAV64061.1 UYSL01021291 VDL77839.1 PDUG01000002 PIC44997.1 UYYA01003809 VDM55829.1 PIC44998.1 UZAD01013320 VDN93953.1 PIC44999.1 CMVM020000306
Proteomes
UP000053240
UP000005204
UP000007266
UP000005203
UP000242457
UP000279307
+ More
UP000007798 UP000001292 UP000008711 UP000007755 UP000000803 UP000002282 UP000075809 UP000000311 UP000008237 UP000268350 UP000192221 UP000008792 UP000008744 UP000001819 UP000002358 UP000001070 UP000009192 UP000053097 UP000078542 UP000053825 UP000078540 UP000007801 UP000235965 UP000092553 UP000005205 UP000078541 UP000053105 UP000078492 UP000183832 UP000009046 UP000245037 UP000000304 UP000030742 UP000014760 UP000046393 UP000232936 UP000038041 UP000274131 UP000024635 UP000095282 UP000038040 UP000274756 UP000218231 UP000008281 UP000216624 UP000008549 UP000008068 UP000237256 UP000216463 UP000085678 UP000001940 UP000031036 UP000050794 UP000267007 UP000005237 UP000095285 UP000038043 UP000271162 UP000230233 UP000050601 UP000267027 UP000095284 UP000038020 UP000278627 UP000024404
UP000007798 UP000001292 UP000008711 UP000007755 UP000000803 UP000002282 UP000075809 UP000000311 UP000008237 UP000268350 UP000192221 UP000008792 UP000008744 UP000001819 UP000002358 UP000001070 UP000009192 UP000053097 UP000078542 UP000053825 UP000078540 UP000007801 UP000235965 UP000092553 UP000005205 UP000078541 UP000053105 UP000078492 UP000183832 UP000009046 UP000245037 UP000000304 UP000030742 UP000014760 UP000046393 UP000232936 UP000038041 UP000274131 UP000024635 UP000095282 UP000038040 UP000274756 UP000218231 UP000008281 UP000216624 UP000008549 UP000008068 UP000237256 UP000216463 UP000085678 UP000001940 UP000031036 UP000050794 UP000267007 UP000005237 UP000095285 UP000038043 UP000271162 UP000230233 UP000050601 UP000267027 UP000095284 UP000038020 UP000278627 UP000024404
Pfam
Interpro
IPR019568
Rapsyn_myristoylation/link_N
+ More
IPR001841 Znf_RING
IPR013026 TPR-contain_dom
IPR013083 Znf_RING/FYVE/PHD
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR001237 Postsynaptic
IPR023298 ATPase_P-typ_TM_dom_sf
IPR032498 PI3K_P85_iSH2
IPR036860 SH2_dom_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR008936 Rho_GTPase_activation_prot
IPR001757 P_typ_ATPase
IPR000198 RhoGAP_dom
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006539 P-type_ATPase_IV
IPR032631 P-type_ATPase_N
IPR036412 HAD-like_sf
IPR000980 SH2
IPR023214 HAD_sf
IPR032630 P_typ_ATPase_c
IPR018303 ATPase_P-typ_P_site
IPR018000 Neurotransmitter_ion_chnl_CS
IPR006201 Neur_channel
IPR009771 Ribosome_control_1
IPR036734 Neur_chan_lig-bd_sf
IPR001965 Znf_PHD
IPR006202 Neur_chan_lig-bd
IPR006028 GABAA/Glycine_rcpt
IPR006029 Neurotrans-gated_channel_TM
IPR036719 Neuro-gated_channel_TM_sf
IPR018957 Znf_C3HC4_RING-type
IPR026000 Apc5_dom
IPR002219 PE/DAG-bd
IPR001841 Znf_RING
IPR013026 TPR-contain_dom
IPR013083 Znf_RING/FYVE/PHD
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR001237 Postsynaptic
IPR023298 ATPase_P-typ_TM_dom_sf
IPR032498 PI3K_P85_iSH2
IPR036860 SH2_dom_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR008936 Rho_GTPase_activation_prot
IPR001757 P_typ_ATPase
IPR000198 RhoGAP_dom
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006539 P-type_ATPase_IV
IPR032631 P-type_ATPase_N
IPR036412 HAD-like_sf
IPR000980 SH2
IPR023214 HAD_sf
IPR032630 P_typ_ATPase_c
IPR018303 ATPase_P-typ_P_site
IPR018000 Neurotransmitter_ion_chnl_CS
IPR006201 Neur_channel
IPR009771 Ribosome_control_1
IPR036734 Neur_chan_lig-bd_sf
IPR001965 Znf_PHD
IPR006202 Neur_chan_lig-bd
IPR006028 GABAA/Glycine_rcpt
IPR006029 Neurotrans-gated_channel_TM
IPR036719 Neuro-gated_channel_TM_sf
IPR018957 Znf_C3HC4_RING-type
IPR026000 Apc5_dom
IPR002219 PE/DAG-bd
SUPFAM
ProteinModelPortal
A0A0N0PDY7
H9JNS3
A0A2H1VFP7
A0A0C9S063
D2A4B4
A0A088AU77
+ More
A0A2A3EP21 A0A3L8DIN3 W8BUF7 W8CDP2 A0A034WI23 A0A0J9S128 B4NHM1 B4IIX3 B3P9S6 F4WQN1 Q7K4Y8 B4PW15 A0A151WV73 E2A7I6 E2BYH2 A0A3B0KT43 A0A1W4VIM2 B4MEZ9 A0A3B0K4F4 A0A310SFW0 B4H875 Q29CY0 K7IML0 B4JZW5 B4L7B4 A0A026WPI6 A0A195D194 A0A0L7RAU4 A0A0J9S0T6 A0A195BRF3 B3MZZ4 A0A2J7PLY3 A0A0M4ES75 A0A158NRZ9 A0A195ERG6 E9IHY2 A0A0M9A2Y4 A0A195D819 A0A1J1HXJ0 A0A1J1HZI4 E0VSU8 A0A336MFR4 A0A3B0K482 A0A1W4VHV8 A0A0A9XSG1 A0A2P8Y3Y9 B4R2F8 U4UZJ4 T1J5G7 R7UF25 A0A0N5ATC2 A0A2A6CT05 A0A0N4V4A9 A0A2G9V5Q0 A0A016UEW2 F1KZS1 A0A1I7TV07 A0A0N4UKH6 A0A2A2LPU3 E3LQX7 A8XCH4 G0NR17 A0A2A2LPD5 A0A2P4V7M8 A0A261BPL8 A0A1S3H1K5 A0A2A2LPB4 Q09485 A0A2A2LP52 A0A0B2V6M7 A0A183V9A1 A0A2A2JR22 K7H5E4 A0A1I7VLE5 A0A0N4YCI1 A0A2G5UZP1 A0A158PFP0 A0A2G5UZP6 A0A1I7RPI5 A0A0N4TVD9 A0A2G5UZR7 A0A044VIV9
A0A2A3EP21 A0A3L8DIN3 W8BUF7 W8CDP2 A0A034WI23 A0A0J9S128 B4NHM1 B4IIX3 B3P9S6 F4WQN1 Q7K4Y8 B4PW15 A0A151WV73 E2A7I6 E2BYH2 A0A3B0KT43 A0A1W4VIM2 B4MEZ9 A0A3B0K4F4 A0A310SFW0 B4H875 Q29CY0 K7IML0 B4JZW5 B4L7B4 A0A026WPI6 A0A195D194 A0A0L7RAU4 A0A0J9S0T6 A0A195BRF3 B3MZZ4 A0A2J7PLY3 A0A0M4ES75 A0A158NRZ9 A0A195ERG6 E9IHY2 A0A0M9A2Y4 A0A195D819 A0A1J1HXJ0 A0A1J1HZI4 E0VSU8 A0A336MFR4 A0A3B0K482 A0A1W4VHV8 A0A0A9XSG1 A0A2P8Y3Y9 B4R2F8 U4UZJ4 T1J5G7 R7UF25 A0A0N5ATC2 A0A2A6CT05 A0A0N4V4A9 A0A2G9V5Q0 A0A016UEW2 F1KZS1 A0A1I7TV07 A0A0N4UKH6 A0A2A2LPU3 E3LQX7 A8XCH4 G0NR17 A0A2A2LPD5 A0A2P4V7M8 A0A261BPL8 A0A1S3H1K5 A0A2A2LPB4 Q09485 A0A2A2LP52 A0A0B2V6M7 A0A183V9A1 A0A2A2JR22 K7H5E4 A0A1I7VLE5 A0A0N4YCI1 A0A2G5UZP1 A0A158PFP0 A0A2G5UZP6 A0A1I7RPI5 A0A0N4TVD9 A0A2G5UZR7 A0A044VIV9
PDB
5A7D
E-value=0.0127467,
Score=91
Ontologies
GO
Topology
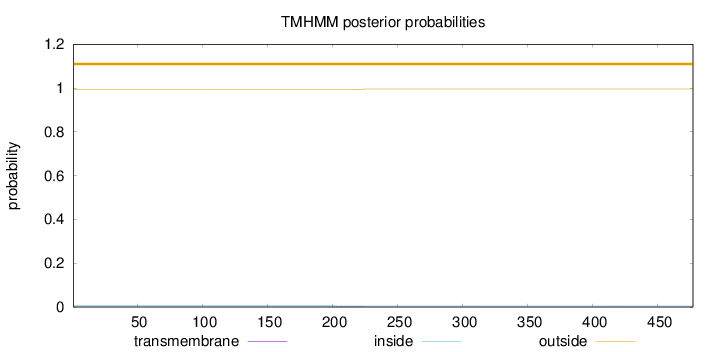
Length:
477
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0453100000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00597
outside
1 - 477
Population Genetic Test Statistics
Pi
18.032183
Theta
17.294459
Tajima's D
-1.159722
CLR
107.295977
CSRT
0.107644617769112
Interpretation
Uncertain
