Gene
KWMTBOMO14160 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011412
Annotation
isocitrate_dehydrogenase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.561
Sequence
CDS
ATGGCTGTGCGTTTGTTAACTAAATTGAAGGTTTTGCCTGATGTGCGCGGGTCTGTGAATCTCAGCTCGAAAGCAGCGCCCGCTACATTATCGGATTTCGATGTCCAGCACAAGACTCCCGTTATCAGGAAACAGAAGCTGATTCCTAAGGCGCAATACGGCGGTCGTCACGCGGTGACCATGCTCCCTGGAGGCGGTATCGGTCCTGAGTGCATGGGATACGTTCGCGACATATTCAAATACATCGGTGCTCCTATTGACTTCGAAGTGGTGGACATTGACCCAACGATGGACAATGATGATGATGTCCAATATGCTATAACGACCATTAAGAGGAACGGTGTGGGGCTAAAGGGCAACATTGAAACCAAAAGTGAGGCAGCCTATGTGACGTCACGCAATGTGGCTCTCAGAAACGAACTGGACATGTATGCTTACATACTGAACTGTAAATCTTACCCTGGCGTTGCGACCAGACATAAGGACATTGATGTTGTTATTATCAGACAGAACACAGAAGGTGAATACGCTATGTTGGAACACGAATCCGTGAATGGTGTGGTCGAGTCAATGAAAGTGGTGACAGCTGACAACTCGGAGAGGGTGGCCAGGTTCGCCTTCGAATTCGCCAAGAAAAACGGCAGGAAGAAGGTAACAACGGTTCACAAAGCCAACATCATGAAGTTATCCGATGGCTTATTCTTGGAGACGTCACGTCGGCTCGCCAAGGAGTACCCGGACATAGAACACAACGATATGATCATTGACAACTGTTGCATGCAGCTCGTCGCAAAGCCACACCAGTTTGATGTTATGCTGATGACTAATCTGTACGGGTCCATTGTCTCGAACGTGGTCTGCGGGCTGTTGGGCGGAGCCGGCCTGCTCTCCGGTAGGAACTACGGCGACCACTACGCTGTATTCGAACCTGGCACTAGGAACACAGGTACCGCCATCGCCGGGAAGAACATTGCCAATCCGATCGCCATGATCAACGCCTCCATAGACATGCTGGAGCATTTGGGCCATCAGTTCCACGCGTCGCTACTCCGCCGCGCCATAAACAAGACGATCAATGTGGACCGCGTACTCACCCCGGATTTGGGCGGATCTAATACCTCCACAGATGTCGTGGGCGCCATCTTACAAAATGTCGGGCGCTGCTAG
Protein
MAVRLLTKLKVLPDVRGSVNLSSKAAPATLSDFDVQHKTPVIRKQKLIPKAQYGGRHAVTMLPGGGIGPECMGYVRDIFKYIGAPIDFEVVDIDPTMDNDDDVQYAITTIKRNGVGLKGNIETKSEAAYVTSRNVALRNELDMYAYILNCKSYPGVATRHKDIDVVIIRQNTEGEYAMLEHESVNGVVESMKVVTADNSERVARFAFEFAKKNGRKKVTTVHKANIMKLSDGLFLETSRRLAKEYPDIEHNDMIIDNCCMQLVAKPHQFDVMLMTNLYGSIVSNVVCGLLGGAGLLSGRNYGDHYAVFEPGTRNTGTAIAGKNIANPIAMINASIDMLEHLGHQFHASLLRRAINKTINVDRVLTPDLGGSNTSTDVVGAILQNVGRC
Summary
Uniprot
H9JPF8
D2Y061
A0A3S2PKZ9
A0A2A4IXR8
A0A212FD56
A0A2W1BDA7
+ More
A0A0N1IPU9 A0A194PUI8 J3JXV8 A0A2J7QPR1 A0A2J7QPP0 V5G5L5 V5GMP4 D6WKK8 A0A1B6EP28 A0A1W4XJK9 A0A1B6M9F3 A0A1B6LLX4 A0A1B6GER4 A0A0A9Z381 A0A0A9XEF1 A0A0K8SEL1 A0A0T6B0Y2 A0A1Y1LHK1 A0A224XRA7 A0A023F9I4 A0A069DSX8 A0A182XWJ8 A0A182M7F3 A0A0J7KFU7 A0A182RFW2 A0A182JY75 A0A182VYC3 A0A084VR08 U5EIY4 A0A182TTJ8 A0A182IYP5 A0A182LQM4 A0A182NMU7 A0A182HZL9 A0A1B6CXN1 Q7QK78 F5HIU6 A0A182WZH2 A0A182V8Y5 A0A182P4G8 A0A2M4A980 A0A2M4BR48 A0A182Q0M5 A0A2M3ZGD6 W5J9R2 A0A182F8A8 A0A1B6DSN8 A0A2M4A8S1 A0A2M4BQN4 A0A195C7W4 A0A336MPM4 A0A0P4VIG3 A0A336MYW2 A0A336MNC9 R4G839 A0A158NU28 A0A336MM72 Q0IEC9 A0A151WZF3 T1DPW9 Q0IED1 Q0IED0 Q0IEC8 A0A195DJK4 A0A182H9M6 A0A195FJX2 E2BYS0 A0A023ER54 E1ZZ09 A0A3L8DW09 B0WQY4 A0A151I4K5 A0A088AKC5 A0A1L8E0A2 A0A1L8E034 A0A1Q3FJE5 A0A1Q3FJS5 A0A0C9Q5R0 A0A0K8TU98 A0A1L8E0A5 E9IRS5 A0A0Q9XCE3 B4K4V5 K7J3R3 I3RSE2 B4M4N6 A0A0Q9WC52 A8JRC1 A0A0N0BH85 A8JRB8 A0A0R1EBV6 Q8MT18 B3P6G3 B4PTH6
A0A0N1IPU9 A0A194PUI8 J3JXV8 A0A2J7QPR1 A0A2J7QPP0 V5G5L5 V5GMP4 D6WKK8 A0A1B6EP28 A0A1W4XJK9 A0A1B6M9F3 A0A1B6LLX4 A0A1B6GER4 A0A0A9Z381 A0A0A9XEF1 A0A0K8SEL1 A0A0T6B0Y2 A0A1Y1LHK1 A0A224XRA7 A0A023F9I4 A0A069DSX8 A0A182XWJ8 A0A182M7F3 A0A0J7KFU7 A0A182RFW2 A0A182JY75 A0A182VYC3 A0A084VR08 U5EIY4 A0A182TTJ8 A0A182IYP5 A0A182LQM4 A0A182NMU7 A0A182HZL9 A0A1B6CXN1 Q7QK78 F5HIU6 A0A182WZH2 A0A182V8Y5 A0A182P4G8 A0A2M4A980 A0A2M4BR48 A0A182Q0M5 A0A2M3ZGD6 W5J9R2 A0A182F8A8 A0A1B6DSN8 A0A2M4A8S1 A0A2M4BQN4 A0A195C7W4 A0A336MPM4 A0A0P4VIG3 A0A336MYW2 A0A336MNC9 R4G839 A0A158NU28 A0A336MM72 Q0IEC9 A0A151WZF3 T1DPW9 Q0IED1 Q0IED0 Q0IEC8 A0A195DJK4 A0A182H9M6 A0A195FJX2 E2BYS0 A0A023ER54 E1ZZ09 A0A3L8DW09 B0WQY4 A0A151I4K5 A0A088AKC5 A0A1L8E0A2 A0A1L8E034 A0A1Q3FJE5 A0A1Q3FJS5 A0A0C9Q5R0 A0A0K8TU98 A0A1L8E0A5 E9IRS5 A0A0Q9XCE3 B4K4V5 K7J3R3 I3RSE2 B4M4N6 A0A0Q9WC52 A8JRC1 A0A0N0BH85 A8JRB8 A0A0R1EBV6 Q8MT18 B3P6G3 B4PTH6
Pubmed
19121390
22118469
28756777
26354079
22516182
23537049
+ More
18362917 19820115 25401762 26823975 28004739 25474469 26334808 25244985 24438588 20966253 12364791 14747013 17210077 20920257 23761445 27129103 21347285 17510324 26483478 20798317 24945155 30249741 26369729 21282665 17994087 20075255 22328714 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
18362917 19820115 25401762 26823975 28004739 25474469 26334808 25244985 24438588 20966253 12364791 14747013 17210077 20920257 23761445 27129103 21347285 17510324 26483478 20798317 24945155 30249741 26369729 21282665 17994087 20075255 22328714 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
BABH01010020
GU270852
ADB27114.1
RSAL01000005
RVE54421.1
NWSH01004998
+ More
PCG64511.1 AGBW02009108 OWR51660.1 KZ150337 PZC71317.1 KQ460036 KPJ18417.1 KQ459593 KPI96414.1 APGK01024794 BT128080 KB740562 KB632399 AEE63041.1 ENN80293.1 ERL94805.1 NEVH01012088 PNF30553.1 PNF30552.1 GALX01003096 JAB65370.1 GALX01003097 JAB65369.1 KQ971342 EFA04017.1 GECZ01030088 JAS39681.1 GEBQ01007429 JAT32548.1 GEBQ01015311 JAT24666.1 GECZ01025032 GECZ01008976 JAS44737.1 JAS60793.1 GBHO01017690 GBHO01005273 GBRD01014477 GDHC01008502 JAG25914.1 JAG38331.1 JAG51349.1 JAQ10127.1 GBHO01025220 JAG18384.1 GBRD01014094 JAG51732.1 LJIG01016290 KRT81046.1 GEZM01055140 GEZM01055139 JAV73114.1 GFTR01005885 JAW10541.1 GBBI01001134 JAC17578.1 GBGD01001691 JAC87198.1 AXCM01009352 LBMM01008125 KMQ89124.1 ATLV01015393 KE525011 KFB40402.1 GANO01002447 JAB57424.1 APCN01001986 GEDC01019213 JAS18085.1 AAAB01008799 EAA03807.5 EGK96207.1 GGFK01003971 MBW37292.1 GGFJ01006333 MBW55474.1 AXCN02001558 GGFM01006828 MBW27579.1 ADMH02001967 ETN60148.1 GEDC01018701 GEDC01009311 GEDC01008620 JAS18597.1 JAS27987.1 JAS28678.1 GGFK01003848 MBW37169.1 GGFJ01006228 MBW55369.1 KQ978143 KYM96740.1 UFQT01001589 SSX31079.1 GDKW01002010 JAI54585.1 SSX31078.1 SSX31081.1 ACPB03011537 GAHY01001642 JAA75868.1 ADTU01026248 SSX31080.1 CH477695 EAT37170.1 KQ982638 KYQ53280.1 GAMD01002964 JAA98626.1 EAT37169.1 EAT37171.1 EAT37168.1 KQ980794 KYN13075.1 JXUM01121172 KQ566470 KXJ70107.1 KQ981522 KYN40656.1 GL451506 EFN79170.1 GAPW01001963 JAC11635.1 GL435250 EFN73576.1 QOIP01000004 RLU23958.1 DS232047 EDS33071.1 KQ976445 KYM86036.1 GFDF01001980 JAV12104.1 GFDF01001981 JAV12103.1 GFDL01007305 JAV27740.1 GFDL01007302 JAV27743.1 GBYB01009433 JAG79200.1 GDAI01000083 JAI17520.1 GFDF01001975 JAV12109.1 GL765283 EFZ16684.1 CH933806 KRG01943.1 EDW16108.2 JQ611724 AFK29227.1 CH940652 EDW59597.2 KRF78948.1 AE014297 ABW08764.1 KQ435759 KOX75664.1 BT082126 ABW08763.1 ACQ57833.1 CM000160 KRK04455.1 AY118436 AAF56494.2 AAM48465.1 CH954182 EDV53633.1 EDW98716.1
PCG64511.1 AGBW02009108 OWR51660.1 KZ150337 PZC71317.1 KQ460036 KPJ18417.1 KQ459593 KPI96414.1 APGK01024794 BT128080 KB740562 KB632399 AEE63041.1 ENN80293.1 ERL94805.1 NEVH01012088 PNF30553.1 PNF30552.1 GALX01003096 JAB65370.1 GALX01003097 JAB65369.1 KQ971342 EFA04017.1 GECZ01030088 JAS39681.1 GEBQ01007429 JAT32548.1 GEBQ01015311 JAT24666.1 GECZ01025032 GECZ01008976 JAS44737.1 JAS60793.1 GBHO01017690 GBHO01005273 GBRD01014477 GDHC01008502 JAG25914.1 JAG38331.1 JAG51349.1 JAQ10127.1 GBHO01025220 JAG18384.1 GBRD01014094 JAG51732.1 LJIG01016290 KRT81046.1 GEZM01055140 GEZM01055139 JAV73114.1 GFTR01005885 JAW10541.1 GBBI01001134 JAC17578.1 GBGD01001691 JAC87198.1 AXCM01009352 LBMM01008125 KMQ89124.1 ATLV01015393 KE525011 KFB40402.1 GANO01002447 JAB57424.1 APCN01001986 GEDC01019213 JAS18085.1 AAAB01008799 EAA03807.5 EGK96207.1 GGFK01003971 MBW37292.1 GGFJ01006333 MBW55474.1 AXCN02001558 GGFM01006828 MBW27579.1 ADMH02001967 ETN60148.1 GEDC01018701 GEDC01009311 GEDC01008620 JAS18597.1 JAS27987.1 JAS28678.1 GGFK01003848 MBW37169.1 GGFJ01006228 MBW55369.1 KQ978143 KYM96740.1 UFQT01001589 SSX31079.1 GDKW01002010 JAI54585.1 SSX31078.1 SSX31081.1 ACPB03011537 GAHY01001642 JAA75868.1 ADTU01026248 SSX31080.1 CH477695 EAT37170.1 KQ982638 KYQ53280.1 GAMD01002964 JAA98626.1 EAT37169.1 EAT37171.1 EAT37168.1 KQ980794 KYN13075.1 JXUM01121172 KQ566470 KXJ70107.1 KQ981522 KYN40656.1 GL451506 EFN79170.1 GAPW01001963 JAC11635.1 GL435250 EFN73576.1 QOIP01000004 RLU23958.1 DS232047 EDS33071.1 KQ976445 KYM86036.1 GFDF01001980 JAV12104.1 GFDF01001981 JAV12103.1 GFDL01007305 JAV27740.1 GFDL01007302 JAV27743.1 GBYB01009433 JAG79200.1 GDAI01000083 JAI17520.1 GFDF01001975 JAV12109.1 GL765283 EFZ16684.1 CH933806 KRG01943.1 EDW16108.2 JQ611724 AFK29227.1 CH940652 EDW59597.2 KRF78948.1 AE014297 ABW08764.1 KQ435759 KOX75664.1 BT082126 ABW08763.1 ACQ57833.1 CM000160 KRK04455.1 AY118436 AAF56494.2 AAM48465.1 CH954182 EDV53633.1 EDW98716.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000019118 UP000030742 UP000235965 UP000007266 UP000192223 UP000076408 UP000075883 UP000036403 UP000075900 UP000075881 UP000075920 UP000030765 UP000075902 UP000075880 UP000075882 UP000075884 UP000075840 UP000007062 UP000076407 UP000075903 UP000075885 UP000075886 UP000000673 UP000069272 UP000078542 UP000015103 UP000005205 UP000008820 UP000075809 UP000078492 UP000069940 UP000249989 UP000078541 UP000008237 UP000000311 UP000279307 UP000002320 UP000078540 UP000005203 UP000009192 UP000002358 UP000008792 UP000000803 UP000053105 UP000002282 UP000008711
UP000019118 UP000030742 UP000235965 UP000007266 UP000192223 UP000076408 UP000075883 UP000036403 UP000075900 UP000075881 UP000075920 UP000030765 UP000075902 UP000075880 UP000075882 UP000075884 UP000075840 UP000007062 UP000076407 UP000075903 UP000075885 UP000075886 UP000000673 UP000069272 UP000078542 UP000015103 UP000005205 UP000008820 UP000075809 UP000078492 UP000069940 UP000249989 UP000078541 UP000008237 UP000000311 UP000279307 UP000002320 UP000078540 UP000005203 UP000009192 UP000002358 UP000008792 UP000000803 UP000053105 UP000002282 UP000008711
Pfam
PF00180 Iso_dh
ProteinModelPortal
H9JPF8
D2Y061
A0A3S2PKZ9
A0A2A4IXR8
A0A212FD56
A0A2W1BDA7
+ More
A0A0N1IPU9 A0A194PUI8 J3JXV8 A0A2J7QPR1 A0A2J7QPP0 V5G5L5 V5GMP4 D6WKK8 A0A1B6EP28 A0A1W4XJK9 A0A1B6M9F3 A0A1B6LLX4 A0A1B6GER4 A0A0A9Z381 A0A0A9XEF1 A0A0K8SEL1 A0A0T6B0Y2 A0A1Y1LHK1 A0A224XRA7 A0A023F9I4 A0A069DSX8 A0A182XWJ8 A0A182M7F3 A0A0J7KFU7 A0A182RFW2 A0A182JY75 A0A182VYC3 A0A084VR08 U5EIY4 A0A182TTJ8 A0A182IYP5 A0A182LQM4 A0A182NMU7 A0A182HZL9 A0A1B6CXN1 Q7QK78 F5HIU6 A0A182WZH2 A0A182V8Y5 A0A182P4G8 A0A2M4A980 A0A2M4BR48 A0A182Q0M5 A0A2M3ZGD6 W5J9R2 A0A182F8A8 A0A1B6DSN8 A0A2M4A8S1 A0A2M4BQN4 A0A195C7W4 A0A336MPM4 A0A0P4VIG3 A0A336MYW2 A0A336MNC9 R4G839 A0A158NU28 A0A336MM72 Q0IEC9 A0A151WZF3 T1DPW9 Q0IED1 Q0IED0 Q0IEC8 A0A195DJK4 A0A182H9M6 A0A195FJX2 E2BYS0 A0A023ER54 E1ZZ09 A0A3L8DW09 B0WQY4 A0A151I4K5 A0A088AKC5 A0A1L8E0A2 A0A1L8E034 A0A1Q3FJE5 A0A1Q3FJS5 A0A0C9Q5R0 A0A0K8TU98 A0A1L8E0A5 E9IRS5 A0A0Q9XCE3 B4K4V5 K7J3R3 I3RSE2 B4M4N6 A0A0Q9WC52 A8JRC1 A0A0N0BH85 A8JRB8 A0A0R1EBV6 Q8MT18 B3P6G3 B4PTH6
A0A0N1IPU9 A0A194PUI8 J3JXV8 A0A2J7QPR1 A0A2J7QPP0 V5G5L5 V5GMP4 D6WKK8 A0A1B6EP28 A0A1W4XJK9 A0A1B6M9F3 A0A1B6LLX4 A0A1B6GER4 A0A0A9Z381 A0A0A9XEF1 A0A0K8SEL1 A0A0T6B0Y2 A0A1Y1LHK1 A0A224XRA7 A0A023F9I4 A0A069DSX8 A0A182XWJ8 A0A182M7F3 A0A0J7KFU7 A0A182RFW2 A0A182JY75 A0A182VYC3 A0A084VR08 U5EIY4 A0A182TTJ8 A0A182IYP5 A0A182LQM4 A0A182NMU7 A0A182HZL9 A0A1B6CXN1 Q7QK78 F5HIU6 A0A182WZH2 A0A182V8Y5 A0A182P4G8 A0A2M4A980 A0A2M4BR48 A0A182Q0M5 A0A2M3ZGD6 W5J9R2 A0A182F8A8 A0A1B6DSN8 A0A2M4A8S1 A0A2M4BQN4 A0A195C7W4 A0A336MPM4 A0A0P4VIG3 A0A336MYW2 A0A336MNC9 R4G839 A0A158NU28 A0A336MM72 Q0IEC9 A0A151WZF3 T1DPW9 Q0IED1 Q0IED0 Q0IEC8 A0A195DJK4 A0A182H9M6 A0A195FJX2 E2BYS0 A0A023ER54 E1ZZ09 A0A3L8DW09 B0WQY4 A0A151I4K5 A0A088AKC5 A0A1L8E0A2 A0A1L8E034 A0A1Q3FJE5 A0A1Q3FJS5 A0A0C9Q5R0 A0A0K8TU98 A0A1L8E0A5 E9IRS5 A0A0Q9XCE3 B4K4V5 K7J3R3 I3RSE2 B4M4N6 A0A0Q9WC52 A8JRC1 A0A0N0BH85 A8JRB8 A0A0R1EBV6 Q8MT18 B3P6G3 B4PTH6
PDB
5YVT
E-value=2.69676e-96,
Score=898
Ontologies
KEGG
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
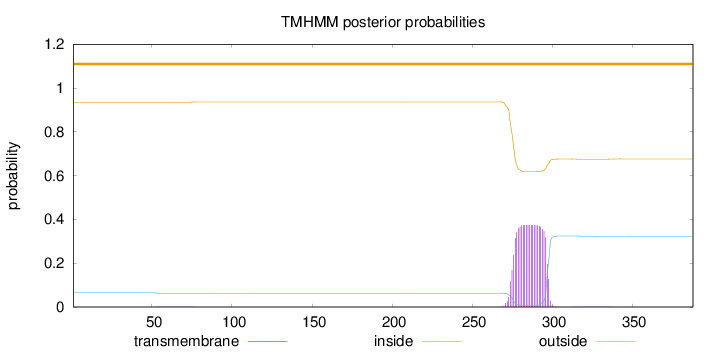
Topology
Length:
388
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.31627999999998
Exp number, first 60 AAs:
0.01306
Total prob of N-in:
0.06480
outside
1 - 388
Population Genetic Test Statistics
Pi
17.558935
Theta
15.763533
Tajima's D
-0.572176
CLR
1.274129
CSRT
0.224438778061097
Interpretation
Uncertain
