Gene
KWMTBOMO14158
Pre Gene Modal
BGIBMGA011411
Annotation
PREDICTED:_CUE_domain-containing_protein_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.279
Sequence
CDS
ATGAGTGCTATAGCGCAACAAGAGTCTTTTGTTAAGGAGAGTCTATTTCAATTTATCATTAAAAACGTTCCAAAAGCAGACTTAAATGTTATTGACGATATAGTCTTGTCTTATGTGATTTCGATACTAGAAGAGGCATCCCAGGATCCCTGCTTCGATGTTGAAGGATTCATCGAGATGATGGCAGCTTACGTACCGGAATTCGCGCACATCGACGCGGGCCTGGTCTGCTCCTGGGTTCTGGAACTGGAAGCCGAGATCTCCCGTCACGAGGAGACCGACTCCAGCAATGGCAACGATGATTCGAGCAGCGATAAAGTGAGCCTTACCCTGCAGAGCCTCTCCGAGATGCTGCCCCCACCACCGAAGAATGATCGCTCCCACACGAACTCCGAGAGTTCCGGCTCTGAGAAGGAGGTGGAAGGAGCCGGGGTGAACATACCCACCTTGGAAGTTCATCAGTGCCAGGTGCTGGCTGAGATGTTTCCTAACGCATGCGCTATGGAGATAAAGCACTGCCTCTCAATTGCCTTTGGCGACGTGGAAAGGGCGGCCGCAACCTTGCTCCACCGCAGGGAGTTGGGCCTCACTCAGTCGGCTCTACACGCTCCGGTCCACCGGACAAAGCCGGTTGACGAACAAGAGGTCAAGCAGAGAATAATCGATCGTTACTCGTACGTGGACGAAAACTCTGGTACGAAGGAGCATAGGCCCTTGGCCCCCAAGATCGAGCCCAAAAAGATGGTCCGGTACAGGGACAATAAGATCGTCTCGTTGAAGGGGGAGCGTTATACCGAGGTCCCCAAATCCGGCGCCGACGAGGAACACCTGAAGAAGCCCAAGAAGCAGCATGCCCCTTAA
Protein
MSAIAQQESFVKESLFQFIIKNVPKADLNVIDDIVLSYVISILEEASQDPCFDVEGFIEMMAAYVPEFAHIDAGLVCSWVLELEAEISRHEETDSSNGNDDSSSDKVSLTLQSLSEMLPPPPKNDRSHTNSESSGSEKEVEGAGVNIPTLEVHQCQVLAEMFPNACAMEIKHCLSIAFGDVERAAATLLHRRELGLTQSALHAPVHRTKPVDEQEVKQRIIDRYSYVDENSGTKEHRPLAPKIEPKKMVRYRDNKIVSLKGERYTEVPKSGADEEHLKKPKKQHAP
Summary
Uniprot
A0A3S2NM31
A0A212F091
A0A0N0PE95
A0A2A4K1Q3
A0A182IM87
A0A1Q3FP17
+ More
W5JKU5 A0A182H2Z4 B0W8G7 Q16HE3 A0A182YAM7 A0A182T672 A0A182K2S7 A0A182QJR2 A0A2W1AZP5 A0A3F2YUP1 A0A182VKK3 A0A182WSF1 Q7PUW5 A0A182HLA2 A0A182NS70 Q8T5K3 A0A182PGR4 A0A182MSW1 A0A182R648 A0A182VZ94 A0A182F7K4 A0A1W4X6R0 A0A1Y1KIX3 W8BAX8 A0A1L8DBW5 T1PL14 A0A1I8PTC3 A0A1I8N7B5 D6WUI4 A0A0K8V976 B4K960 A0A1B0CF35 A0A182TFD7 A0A1A9VER3 A0A1B0F9Q8 A0A1A9ZXN2 A0A067RBD4 A0A1A9XZA2 A0A1A9W3I0 A0A336KXG2 N6TLL8 A0A0A9YP72 A0A023EXJ6 A0A224XNX6 A0A069DYK1 A0A0V0GD88 T1IBP1 A0A1B6K8I9 A0A0C9QKH2 A0A1J1IGA6 A0A1B6MS13 E2AKA3 A0A195AUM7 A0A158NHJ7 A0A0J7KV17 A0A1B6DBI2 A0A195EUK1 F4WVR6 E2B9I8 A0A195E291 A0A026WXF8 A0A0M8ZNB2 A0A0L7QWV0 A0A2A3ELU7 V9IIR7 A0A151WGN7 A0A088AD61 E9J0E7 A0A232ELQ5 A0A310SP41 A0A154PEH8 A0A0P5HGY9 E9G5C1 K7J1Z5 A0A0P5KRF4 A0A195D151 A0A0P5KTN1 A0A0P4X4I8 A0A1Y1KH46 L7M7C0 A0A293LBQ2 A0A2R5LED6 A0A0P5FIQ2 A0A084VI65 T1JF40 A0A1S3JGI2 A0A1B6IT80 A0A1E1X5Z4 A0A1S3JHY3 W8BFW2 A0A210Q326
W5JKU5 A0A182H2Z4 B0W8G7 Q16HE3 A0A182YAM7 A0A182T672 A0A182K2S7 A0A182QJR2 A0A2W1AZP5 A0A3F2YUP1 A0A182VKK3 A0A182WSF1 Q7PUW5 A0A182HLA2 A0A182NS70 Q8T5K3 A0A182PGR4 A0A182MSW1 A0A182R648 A0A182VZ94 A0A182F7K4 A0A1W4X6R0 A0A1Y1KIX3 W8BAX8 A0A1L8DBW5 T1PL14 A0A1I8PTC3 A0A1I8N7B5 D6WUI4 A0A0K8V976 B4K960 A0A1B0CF35 A0A182TFD7 A0A1A9VER3 A0A1B0F9Q8 A0A1A9ZXN2 A0A067RBD4 A0A1A9XZA2 A0A1A9W3I0 A0A336KXG2 N6TLL8 A0A0A9YP72 A0A023EXJ6 A0A224XNX6 A0A069DYK1 A0A0V0GD88 T1IBP1 A0A1B6K8I9 A0A0C9QKH2 A0A1J1IGA6 A0A1B6MS13 E2AKA3 A0A195AUM7 A0A158NHJ7 A0A0J7KV17 A0A1B6DBI2 A0A195EUK1 F4WVR6 E2B9I8 A0A195E291 A0A026WXF8 A0A0M8ZNB2 A0A0L7QWV0 A0A2A3ELU7 V9IIR7 A0A151WGN7 A0A088AD61 E9J0E7 A0A232ELQ5 A0A310SP41 A0A154PEH8 A0A0P5HGY9 E9G5C1 K7J1Z5 A0A0P5KRF4 A0A195D151 A0A0P5KTN1 A0A0P4X4I8 A0A1Y1KH46 L7M7C0 A0A293LBQ2 A0A2R5LED6 A0A0P5FIQ2 A0A084VI65 T1JF40 A0A1S3JGI2 A0A1B6IT80 A0A1E1X5Z4 A0A1S3JHY3 W8BFW2 A0A210Q326
Pubmed
22118469
26354079
20920257
23761445
26483478
17510324
+ More
25244985 28756777 20966253 12364791 14747013 17210077 12060762 28004739 24495485 25315136 18362917 19820115 17994087 24845553 23537049 25401762 26823975 25474469 26334808 20798317 21347285 21719571 24508170 30249741 21282665 28648823 21292972 20075255 25576852 24438588 28503490 28812685
25244985 28756777 20966253 12364791 14747013 17210077 12060762 28004739 24495485 25315136 18362917 19820115 17994087 24845553 23537049 25401762 26823975 25474469 26334808 20798317 21347285 21719571 24508170 30249741 21282665 28648823 21292972 20075255 25576852 24438588 28503490 28812685
EMBL
RSAL01000005
RVE54422.1
AGBW02011165
OWR47114.1
KQ460036
KPJ18416.1
+ More
NWSH01000273 PCG77858.1 GFDL01005705 JAV29340.1 ADMH02001150 ETN63908.1 JXUM01106462 KQ565058 KXJ71403.1 DS231859 EDS39017.1 CH478180 EAT33670.1 AXCN02000267 KZ150560 PZC70532.1 AAAB01008987 EAA00904.3 APCN01000888 AJ439060 CAD27757.1 AXCM01006747 GEZM01087359 JAV58737.1 GAMC01010723 JAB95832.1 GFDF01010145 JAV03939.1 KA648648 AFP63277.1 KQ971357 EFA08382.1 GDHF01028973 GDHF01016795 JAI23341.1 JAI35519.1 CH933806 EDW14473.1 AJWK01009644 CCAG010019856 KK852618 KDR20147.1 UFQS01000635 UFQT01000635 SSX05610.1 SSX25969.1 APGK01031440 APGK01031441 APGK01031442 APGK01031443 KB740809 KB631718 ENN78883.1 ERL85569.1 GBHO01010706 GBRD01005879 GDHC01001577 JAG32898.1 JAG59942.1 JAQ17052.1 GBBI01005031 JAC13681.1 GFTR01004888 JAW11538.1 GBGD01002250 JAC86639.1 GECL01000229 JAP05895.1 ACPB03004975 GECU01000213 JAT07494.1 GBYB01001102 GBYB01001104 JAG70869.1 JAG70871.1 CVRI01000050 CRK99277.1 GEBQ01001247 JAT38730.1 GL440209 EFN66144.1 KQ976737 KYM75943.1 ADTU01015860 LBMM01002956 KMQ94138.1 GEDC01014254 JAS23044.1 KQ981975 KYN31577.1 GL888393 EGI61736.1 GL446556 EFN87621.1 KQ979763 KYN19270.1 KK107069 QOIP01000002 EZA60707.1 RLU25870.1 KQ436010 KOX67577.1 KQ414706 KOC63103.1 KZ288220 PBC32176.1 JR047855 AEY60527.1 KQ983167 KYQ46965.1 GL767462 EFZ13702.1 NNAY01003518 OXU19285.1 KQ759773 OAD62925.1 KQ434889 KZC10275.1 GDIQ01227665 LRGB01000568 JAK24060.1 KZS18049.1 GL732532 EFX85181.1 GDIQ01180773 JAK70952.1 KQ976973 KYN06635.1 GDIQ01179378 JAK72347.1 GDIP01246462 JAI76939.1 GEZM01087361 JAV58736.1 GACK01006030 JAA59004.1 GFWV01000498 MAA25228.1 GGLE01003736 MBY07862.1 GDIQ01256575 JAJ95149.1 ATLV01013301 KE524850 KFB37659.1 JH432141 GECU01017578 JAS90128.1 GFAC01004732 JAT94456.1 GAMC01010722 JAB95833.1 NEDP02005172 OWF43069.1
NWSH01000273 PCG77858.1 GFDL01005705 JAV29340.1 ADMH02001150 ETN63908.1 JXUM01106462 KQ565058 KXJ71403.1 DS231859 EDS39017.1 CH478180 EAT33670.1 AXCN02000267 KZ150560 PZC70532.1 AAAB01008987 EAA00904.3 APCN01000888 AJ439060 CAD27757.1 AXCM01006747 GEZM01087359 JAV58737.1 GAMC01010723 JAB95832.1 GFDF01010145 JAV03939.1 KA648648 AFP63277.1 KQ971357 EFA08382.1 GDHF01028973 GDHF01016795 JAI23341.1 JAI35519.1 CH933806 EDW14473.1 AJWK01009644 CCAG010019856 KK852618 KDR20147.1 UFQS01000635 UFQT01000635 SSX05610.1 SSX25969.1 APGK01031440 APGK01031441 APGK01031442 APGK01031443 KB740809 KB631718 ENN78883.1 ERL85569.1 GBHO01010706 GBRD01005879 GDHC01001577 JAG32898.1 JAG59942.1 JAQ17052.1 GBBI01005031 JAC13681.1 GFTR01004888 JAW11538.1 GBGD01002250 JAC86639.1 GECL01000229 JAP05895.1 ACPB03004975 GECU01000213 JAT07494.1 GBYB01001102 GBYB01001104 JAG70869.1 JAG70871.1 CVRI01000050 CRK99277.1 GEBQ01001247 JAT38730.1 GL440209 EFN66144.1 KQ976737 KYM75943.1 ADTU01015860 LBMM01002956 KMQ94138.1 GEDC01014254 JAS23044.1 KQ981975 KYN31577.1 GL888393 EGI61736.1 GL446556 EFN87621.1 KQ979763 KYN19270.1 KK107069 QOIP01000002 EZA60707.1 RLU25870.1 KQ436010 KOX67577.1 KQ414706 KOC63103.1 KZ288220 PBC32176.1 JR047855 AEY60527.1 KQ983167 KYQ46965.1 GL767462 EFZ13702.1 NNAY01003518 OXU19285.1 KQ759773 OAD62925.1 KQ434889 KZC10275.1 GDIQ01227665 LRGB01000568 JAK24060.1 KZS18049.1 GL732532 EFX85181.1 GDIQ01180773 JAK70952.1 KQ976973 KYN06635.1 GDIQ01179378 JAK72347.1 GDIP01246462 JAI76939.1 GEZM01087361 JAV58736.1 GACK01006030 JAA59004.1 GFWV01000498 MAA25228.1 GGLE01003736 MBY07862.1 GDIQ01256575 JAJ95149.1 ATLV01013301 KE524850 KFB37659.1 JH432141 GECU01017578 JAS90128.1 GFAC01004732 JAT94456.1 GAMC01010722 JAB95833.1 NEDP02005172 OWF43069.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000218220
UP000075880
UP000000673
+ More
UP000069940 UP000249989 UP000002320 UP000008820 UP000076408 UP000075901 UP000075881 UP000075886 UP000075882 UP000075903 UP000076407 UP000007062 UP000075840 UP000075884 UP000075885 UP000075883 UP000075900 UP000075920 UP000069272 UP000192223 UP000095300 UP000095301 UP000007266 UP000009192 UP000092461 UP000075902 UP000078200 UP000092444 UP000092445 UP000027135 UP000092443 UP000091820 UP000019118 UP000030742 UP000015103 UP000183832 UP000000311 UP000078540 UP000005205 UP000036403 UP000078541 UP000007755 UP000008237 UP000078492 UP000053097 UP000279307 UP000053105 UP000053825 UP000242457 UP000075809 UP000005203 UP000215335 UP000076502 UP000076858 UP000000305 UP000002358 UP000078542 UP000030765 UP000085678 UP000242188
UP000069940 UP000249989 UP000002320 UP000008820 UP000076408 UP000075901 UP000075881 UP000075886 UP000075882 UP000075903 UP000076407 UP000007062 UP000075840 UP000075884 UP000075885 UP000075883 UP000075900 UP000075920 UP000069272 UP000192223 UP000095300 UP000095301 UP000007266 UP000009192 UP000092461 UP000075902 UP000078200 UP000092444 UP000092445 UP000027135 UP000092443 UP000091820 UP000019118 UP000030742 UP000015103 UP000183832 UP000000311 UP000078540 UP000005205 UP000036403 UP000078541 UP000007755 UP000008237 UP000078492 UP000053097 UP000279307 UP000053105 UP000053825 UP000242457 UP000075809 UP000005203 UP000215335 UP000076502 UP000076858 UP000000305 UP000002358 UP000078542 UP000030765 UP000085678 UP000242188
Pfam
PF02845 CUE
SUPFAM
SSF46934
SSF46934
ProteinModelPortal
A0A3S2NM31
A0A212F091
A0A0N0PE95
A0A2A4K1Q3
A0A182IM87
A0A1Q3FP17
+ More
W5JKU5 A0A182H2Z4 B0W8G7 Q16HE3 A0A182YAM7 A0A182T672 A0A182K2S7 A0A182QJR2 A0A2W1AZP5 A0A3F2YUP1 A0A182VKK3 A0A182WSF1 Q7PUW5 A0A182HLA2 A0A182NS70 Q8T5K3 A0A182PGR4 A0A182MSW1 A0A182R648 A0A182VZ94 A0A182F7K4 A0A1W4X6R0 A0A1Y1KIX3 W8BAX8 A0A1L8DBW5 T1PL14 A0A1I8PTC3 A0A1I8N7B5 D6WUI4 A0A0K8V976 B4K960 A0A1B0CF35 A0A182TFD7 A0A1A9VER3 A0A1B0F9Q8 A0A1A9ZXN2 A0A067RBD4 A0A1A9XZA2 A0A1A9W3I0 A0A336KXG2 N6TLL8 A0A0A9YP72 A0A023EXJ6 A0A224XNX6 A0A069DYK1 A0A0V0GD88 T1IBP1 A0A1B6K8I9 A0A0C9QKH2 A0A1J1IGA6 A0A1B6MS13 E2AKA3 A0A195AUM7 A0A158NHJ7 A0A0J7KV17 A0A1B6DBI2 A0A195EUK1 F4WVR6 E2B9I8 A0A195E291 A0A026WXF8 A0A0M8ZNB2 A0A0L7QWV0 A0A2A3ELU7 V9IIR7 A0A151WGN7 A0A088AD61 E9J0E7 A0A232ELQ5 A0A310SP41 A0A154PEH8 A0A0P5HGY9 E9G5C1 K7J1Z5 A0A0P5KRF4 A0A195D151 A0A0P5KTN1 A0A0P4X4I8 A0A1Y1KH46 L7M7C0 A0A293LBQ2 A0A2R5LED6 A0A0P5FIQ2 A0A084VI65 T1JF40 A0A1S3JGI2 A0A1B6IT80 A0A1E1X5Z4 A0A1S3JHY3 W8BFW2 A0A210Q326
W5JKU5 A0A182H2Z4 B0W8G7 Q16HE3 A0A182YAM7 A0A182T672 A0A182K2S7 A0A182QJR2 A0A2W1AZP5 A0A3F2YUP1 A0A182VKK3 A0A182WSF1 Q7PUW5 A0A182HLA2 A0A182NS70 Q8T5K3 A0A182PGR4 A0A182MSW1 A0A182R648 A0A182VZ94 A0A182F7K4 A0A1W4X6R0 A0A1Y1KIX3 W8BAX8 A0A1L8DBW5 T1PL14 A0A1I8PTC3 A0A1I8N7B5 D6WUI4 A0A0K8V976 B4K960 A0A1B0CF35 A0A182TFD7 A0A1A9VER3 A0A1B0F9Q8 A0A1A9ZXN2 A0A067RBD4 A0A1A9XZA2 A0A1A9W3I0 A0A336KXG2 N6TLL8 A0A0A9YP72 A0A023EXJ6 A0A224XNX6 A0A069DYK1 A0A0V0GD88 T1IBP1 A0A1B6K8I9 A0A0C9QKH2 A0A1J1IGA6 A0A1B6MS13 E2AKA3 A0A195AUM7 A0A158NHJ7 A0A0J7KV17 A0A1B6DBI2 A0A195EUK1 F4WVR6 E2B9I8 A0A195E291 A0A026WXF8 A0A0M8ZNB2 A0A0L7QWV0 A0A2A3ELU7 V9IIR7 A0A151WGN7 A0A088AD61 E9J0E7 A0A232ELQ5 A0A310SP41 A0A154PEH8 A0A0P5HGY9 E9G5C1 K7J1Z5 A0A0P5KRF4 A0A195D151 A0A0P5KTN1 A0A0P4X4I8 A0A1Y1KH46 L7M7C0 A0A293LBQ2 A0A2R5LED6 A0A0P5FIQ2 A0A084VI65 T1JF40 A0A1S3JGI2 A0A1B6IT80 A0A1E1X5Z4 A0A1S3JHY3 W8BFW2 A0A210Q326
Ontologies
GO
PANTHER
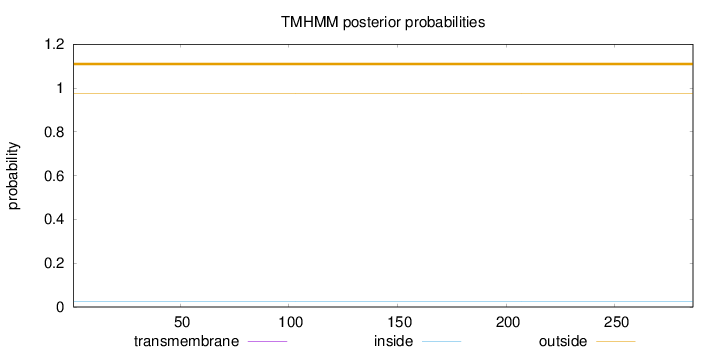
Topology
Length:
286
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00194
Exp number, first 60 AAs:
0.00027
Total prob of N-in:
0.02647
outside
1 - 286
Population Genetic Test Statistics
Pi
17.91779
Theta
14.323925
Tajima's D
0.368808
CLR
0.149415
CSRT
0.484675766211689
Interpretation
Uncertain
